Abstract
Eukaryotic cells devoid of mitochondrial DNA (ρ0 cells) were originally generated under artificial growth conditions utilizing ethidium bromide. The chemical is known to intercalate preferentially with the mitochondrial double-stranded DNA thereby interfering with enzymes of the replication machinery. ρ0 cell lines are highly valuable tools to study human mitochondrial disorders because they can be utilized in cytoplasmic transfer experiments. However, mutagenic effects of ethidium bromide onto the nuclear DNA cannot be excluded. To foreclose this mutagenic character during the development of ρ0 cell lines, we developed an extremely mild, reliable and timesaving method to generate ρ0 cell lines within 3–5 days based on an enzymatic approach. Utilizing the genes for the restriction endonuclease EcoRI and the fluorescent protein EGFP that were fused to a mitochondrial targeting sequence, we developed a CMV-driven expression vector that allowed the temporal expression of the resulting fusion enzyme in eukaryotic cells. Applied on the human cell line 143B.TK− the active protein localized to mitochondria and induced the complete destruction of endogenous mtDNA. Mouse and rat ρ0 cell lines were also successfully created with this approach. Furthermore, the newly established 143B.TK− ρ0 cell line was characterized in great detail thereby releasing interesting insights into the morphology and ultra structure of human ρ0 mitochondria.
INTRODUCTION
Mitochondria are organelles that can be found in most eukaryotic cells. Mitochondria harbour critical biochemical processes, such as the Krebs cycle or the aerobic energy supply of the cell. More recently, however, it was shown that they are key players in the ageing process and the programmed cell death. One factor closely linked to all these functions is the genome of the organelle, the mitochondrial DNA. Numerous neurological and neuromuscular diseases presenting with a variety of symptoms have been associated with mutations of the mitochondrial genome (1). Mitochondrial encephalopathies for example are a class of diseases which result from dysfunction of the mitochondria's oxidative phosphorylation system (OXPHOS). Under regular conditions, this system takes responsibility for cellular respiration and energy production. The associated diseases exhibit disorders manifesting in tissues with high aerobic metabolic demands (e.g. brain, skeletal muscle, heart). The OXPHOS involves five enzyme complexes that assemble from subunits encoded by the nuclear DNA (nDNA) and the mitochondrial DNA (mtDNA). The mitochondrial genome is organized in a circular fashion, encompassing 16 569 bp (according to 2) and encoding 13 polypeptides involved in OXPHOS, a set of 22 essential tRNAs as well as the large (16 S) and small (12 S) ribosomal RNA required for mitochondrial translation. Although the mtDNA was the first eukaryotic genome completely sequenced, almost nothing is known about the processes of how nucleus and mitochondria interact, of how mtDNA and nDNA gene expression is coordinated or how mtDNA is maintained within the cell (a process strictly driven by nuclear factors). Resulting from this lack of knowledge, very little is known about the molecular mechanisms leading to human diseases as a result of mtDNA damage. This is due, in part, to our inability to study regulatory and developmental mitochondrial processes under experimental conditions.
One effort to overcome these difficulties was an experiment carried out by King and Attardi (3). A human osteosarcoma cell line (143B.TK−, derived from an osteosarcoma) was treated over 4–6 weeks with a low dosage of ethidium bromide. It is known from yeast experiments (4–6) that this reagent interacts with mtDNA to form complexes that interfere with DNA replication. After a series of cell divisions, the endogenous mtDNA is lost, while nuclear DNA is maintained. Since the loss of mtDNA extinguishes the oxidative phosphorylation system after a few days, cells devoid of endogenous mtDNA (so called ρ0 cells) need supplementation with nutrients to sustain viability. This is achieved by adding pyruvate and uridine to the growth medium so that the energy demand of the cell is satisfied additionally to the possibility of generating pyrimidines in spite of the inhibition of dihydroorotate dehydrogenase (DHODH) due to loss of respiratory chain electron transfer.
Additionally, other reagents were tested to generate ρ0 cells that interfere with mtDNA replication (ditercalinium, ddC, etc.) (7–10). However, all chemicals used presented severe disadvantages, such as mutagenic effects or the induction of mdr (multi-drug resistance) family gene expression so that the inhibitory effect on mtDNA replication was abolished. Moreover, the major limitation of these methods is their inapplicability to different cell lines. For example, despite anecdotal evidence of many attempts, there are no published accounts of rat ρ0 cell lines produced by these methods.
We present a strategy based on a restriction endonuclease targeted to the matrix of mitochondria that allows the destruction of all endogenous mtDNA. The DNA is cleaved by the restriction endonuclease and endogenous nucleases act to fully disintegrate the mtDNA. This strategy was realized by selecting the gene for the restriction endonuclease EcoRI that is known to cleave human mtDNA 3–5 times. By fusing the EcoRI gene to a mitochondrial targeting sequence derived from the gene of subunit VIII of human cytochrome c oxidase and to the gene of EGFP (enhanced green fluorescent protein) as optical marker, mitochondrial localization was achieved. After transfection, selection with either geneticine or FACS analyses and growth of a cell clone designated as 143B.TK−K7, the loss of all endogenous mitochondrial DNA was confirmed by metabolic testing, PCR and Southern blot analyses. Additional comparative studies were carried out to characterize the proliferative, metabolic and morphological changes due to a ρ0 state of these cells. Therefore a 143B.TK− ρ0 cell line generated by incubation with low doses of ethidium bromide and the newly developed 143B.TK−K7 cell clone were compared to wild-type cells for growth rate, glucose consumption as well as lactate and proton production rate.
MATERIALS AND METHODS
Cloning
Cloning was performed according to standard procedures, and all polymerase chain reaction (PCR) products were verified by sequencing.
The plasmid pAN4 [a kind gift of A. Kiss and constructed by P. Modrich and co-workers (11)] coding for the EcoRI gene was used as template in a PCR with following primers: EcoRIR-001-FOR (2) 5′-catggacgagctgtacaagatgtctaataaaaaac-3′, which adds a short sequence complementary to the 3′-end of the EGFP gene and EcoRIR-834-REV (2) 5′-ggccaaatcacttagatgtaagctgttcaaac-3′ which generates a NotI-restriction site. The EGFP gene from the plasmid pEGFP-Mito (Clontech) was amplified with the primers pEGFP-Mito-0597-FOR 5′-ggccaaatgtccgtcctgacg-3′ which generates a NotI-restriction site and pEGFP-Mito-1421-REV 5′-cttgtacagctcgtccatgccg-3′ which amplifies the EGFP without the stop-codon. Both fragments were utilized in a recombinant PCR and the phosphorylated product was sub-cloned in the PvuII-linearized vector pTRE2hyg. The fusion gene product consisting of EGFP- and EcoRI gene was cloned with AgeI and NotI into pEGFP-Mito resulting in the vector pMEE-con.
Cell culture, transfection and mitochondrial staining
Human osteosarcoma cells 143B.TK− (ATCC CRL-8303) were cultured under standard conditions in Dulbecco's modified Eagle's medium (high glucose) with GlutaMAX (Invitrogen) supplemented with 10% fetal calf serum, 1% bromdeoxyuridine, 100 U penicillin and 100 µg/ml streptomycin. The ρ0 cell line referred to as 143B.TK− ρ0 was generated by incubation of 143B.TK− with low doses of ethidium bromide. ρ0 cell lines of the parental 143B.TK− and cells after transfection procedure were maintained as indicated with additional supplementation with 100 µg/ml pyruvate and 50 µg/ml uridine.
Transient transfections of 143B.TK− and 143B.DsRed1-Mito (143B.TK− cells stably expressing mitochondrially targeted DsRed1 for co-localization studies with pMEE-con) were performed using Effectene (Qiagen) according to manufacturer's conditions. Geneticin selection with 700 µg/ml was carried out 24 h after transfection for 5 days. An isolated clone named as 143B.TK−K7 was aliquoted and metabolically tested for existing oxidative phosphorylation with DMEM (high glucose) with GlutaMAX, 10% dialysed FCS (3), with 1% bromdeoxyuridine, 100 U penicillin and 100 µg/ml streptomycin without pyruvate and uridine for 30 days.
Two other cell lines were used to extend the applicability of our approach to rodent cells: the mouse line LMTK− (ATCC CCL-1.3), and the rat line NRK52E (ATCC CRL-1571).
MitoTracker CMXRos (Molecular Probes) was used to label mitochondria according to manufacturer's protocol at a concentration of 100 nM.
FACS analyses
Samples of transfected cells (2 × 106 cells/ml in 0.05% BSA in PBS) were analysed on a flow cytometer and its software (FACSCAN, FACS Vantage SE, Cell Quest Software, BD Biosciences). Acquisition of data was performed by gating more than 100 000 cells expressing the mitochondrial targeted restriction endonuclease fused to EGFP with the flow cytometer.
Characterization of clones by PCR
To verify the loss of all endogenous mtDNA in the resulting 143B.TK−K7 cell clone the genomic DNA was isolated and used as a template in a PCR. Controls were accomplished by using the genomic DNA isolated from the 143B.TK− wild type and a corresponding ρ0 cell-line generated by incubation with ethidium bromide as described before. The amplified region of the mtDNA does not contain EcoRI sites. Moreover, the primers selected anneal within the D-loop area encompassing the origin (OH) of mtDNA replication. Persisting mtDNA molecules would be expected to contain this region to further possess DNA replication fidelity. The following primers binding in the mtDNA region were utilized: 15 501-FOR 5′-acccagacaattataccctagc-3′ and 630 REV 5′-gagcccgtctaaacattttcaatg-3′. The control-PCR was performed with primers that amplify the human histone H1 gene (Histon H1 5′ 5′-atgagctcatgaccgagaattccacgtccg-3′ and Histon H1 3′ 5′-atcccgggcaaacttcttcttgcc-3′). The annealing temperature was 55°C for 35 cycles with an elongation time of 1 min.
Similar analyses were carried out on rat and mouse DNA extracted from LMTK− and NRK52E cell lines. Primers utilized for mtDNA amplification were Rnor-15525-For 5′-ctataaattcacaacaacatgtc-3′ and Rnor-1214-Rev 5′-ttaagctctctccccccgtatccg-3′ (rat cell line) and Mmus-16033-For 5′-ctaattattcatgcttgttagac-3′ and Mmus-1174-Rev 5′-tccaatacttttagtaggataaat-3′ (mouse cell line). Histones were amplified utilizing primers Rnor-HistonH3.3-001-For 5′-atggcccgaaccaagcagacc-3′ and Rnor-HistonH3.3-539-Rev 5′-ttaagctctctccccccgtatccg-3′ (rat cell line), and Mmus-Histon 3a-001-For 5′-atggctcgtacaaagcagactgcc-3′ and Mmus-Histon3a-411-Rev 5′-ttaagcacgttctccgcgtatgc-3′ (mouse cell line).
Southern blot analysis
Total DNA (15 µg) extracted from cells was digested with the restriction endonucleases BamHI or PvuII. Restricted fragments were separated on a 0.6% agarose gel, transferred onto a nylon membrane and hybridized with a digoxygenin-dUTP-labelled human mtDNA probe amplified with mtDNA primers annealing to nucleotides 4831–4846 (For) and 5628–5651 (Rev). The membrane was washed, and the fragments were detected with the DIG Nucleic Acid Detection Kit (Roche Applied Sciences) according to the manufacturer's instructions. Chemoluminescence of the fragments was analysed with a bioimaging system.
Metabolite analyses of culture medium
In a 35-mm dish, 4 × 105 cells were seeded and incubated in 2.5 ml culture medium supplemented with 100 µg/ml pyruvate and 50 µg/ml uridine under standard conditions. After 24, 40, 48 and 62 h cultivation time the cell number was quantified, the pH value analysed and the glucose and lactate concentrations determined on a Hitachi 917 clinical chemistry analyser (Roche Diagnostics). The pH value was converted into proton concentration in medium by the equation proton conc. (mol/l) = 10–pH, standardized additionally to all other data to 106 cells and analysed graphically. The indicated values are averages of four individual measurements or eight independent measurements (pH value of ρ0 cells), respectively. Standard deviations are depicted as error bars.
Confocal microscopy
Living cells cultured on glass bottom dishes (MatTek Corporation Ashland) were observed with the inverted confocal laser scanning microscope TCS SP5 (Leica Microsystems). To avoid a cross talk in excitation of multiple stained compounds a sequential scanning mode was used in confocal microscopy exclusively.
Images were acquired with photo multipliers and micrographs were processed and analysed with the software Leica Application Suite Advanced Fluorescence 1.5.1 and Adobe Photoshop CS.
Electron microscopy
Cells grown on cover slips were fixed with a solution of 2.5% glutaraldehyde, 2% formaldehyde (made from paraformaldehyde) in 100 mM cacodylate buffer, pH 7.4 for 1.5 h at 4°C, washed twice with cacodylate buffer, followed by a fixation with 2% osmium tetroxide in 50 mM cacodylate buffer (pH 7.4). Specimens were washed twice with distilled water and stained over night with aqueous 0.5% uranyl acetate at 4°C. Cells were dehydrated and flat embedded in Epon 812. Ultra-thin sections were analysed with a Zeiss EM10 (Oberkochen, Germany). Negatives were digitized by scanning and processed with Adobe Photoshop CS.
RESULTS
Generation of ρ0 cells utilizing a mitochondrial targeted restriction endonuclease
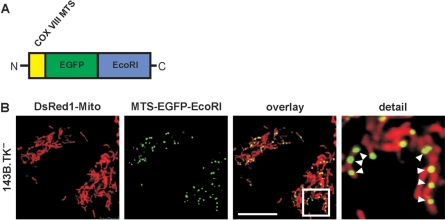
The aim of this work was to develop a system for generating cells devoid of any endogenous mitochondrial DNA without using mutagenic substances like ethidium bromide, ditercalinum or 2′3′-dideoxycytidin. The basis of this system was the vector pMEE-con that after transfection in cells constitutively expresses the restriction endonuclease EcoRI as functional unit. EcoRI is known to cleave human mtDNA 3–5 times depending on the cells’ ethnic background. Laboratory mouse mtDNA also has three EcoRI sites, while Rattus norvegicus, the source of the NRK52E cell line, has seven sites. EcoRI was fused to a mitochondrial targeting peptide derived from the human cytochrome c oxidase subunit 8 (COX VIII). Furthermore, this construct was fused to the enhanced green fluorescent protein EGFP (Figure 1A) on the one hand to observe the expression in the cells (Figure 1B) and to have a selection marker suitable for FACS analysis on the other hand so that transfected cells can be selected to purity. Confocal fluorescence microscopy showed that the polypeptide was expressed and presented a mitochondrial localization in punctate structures presumably in close contact to mtDNA in nucleoids (Figure 1B). After selection of transient transfected cells with the antibiotic geneticin for 5 days or alternatively by FACS selection, a clone of 143B.TK− could be isolated. This cell clone designated as 143B.TK−K7 was metabolically tested for its dependency on pyruvate and uridine. The cells exhibited only poor proliferation during cultivation in medium without pyruvate und uridine and finally died, strongly suggesting a ρ0-state of the cell line.
Figure 1.
Construction and expression of the mitochondrially targeted EcoRI. A DNA fragment coding for the COX VIII targeting sequence (MTS) was added to the 5′ end of the restriction endonuclease gene EcoRI and as optical marker the gene for the enhanced green fluorescent protein EGFP was fused to the 3′ end. The construct was sub-cloned into the vector pTRE2hyg and re-cloned with AgeI and NotI into pEGFP-Mito resulting in the vector pMEE-con with a constitutive CMV promoter (A) The final construct was transfected in 143B.DsRed1-Mito cells. Twenty-four hours after transfection the mitochondrial localization of the EcoRI fusion protein in punctate structures (white arrowheads) was confirmed by co-localization with DsRed1-Mito with confocal fluorescence microscopy (B) The calibration mark corresponds to 10 µm.
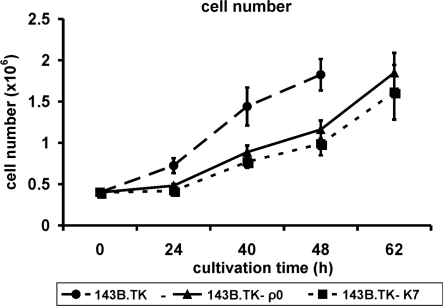
The analyses of cell proliferation during 62 h of cultivation time (Figure 2) showed an obvious decrease in proliferation rate of the 143B.TK−K7 cells (dotted lines) in contrast to the wild type (dashed lines). This is consistent with observations seen in a ρ0 cell line referred to as 143B.TK− ρ0 generated by incubation with low doses of ethidium bromide (solid line). Both ρ0 cell lines exhibit a similar genetic background. The figure indicates that the 143B.TK−K7 cells possess a similar growth rate as the 143B.TK− ρ0 cells.
Figure 2.
Graphic documentation of the proliferation rate of the cell lines 143B.TK−K7 (dotted line), 143B.TK− ρ0 (solid line) and 143B.TK− (dashed line) at proceeding cultivation times. The indicated values are averages of four individual measurements. The standard deviations are depicted as error bars. The ρ0 cells exhibit a decelerated proliferation rate compared to the wild type. Time scale represents the time intervals proceeded after cell seeding.
Confirmation of ρ0 state
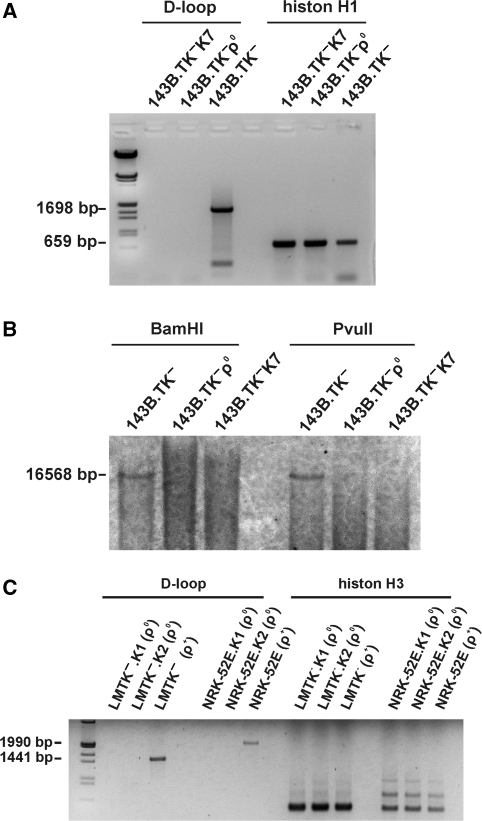
Additional confirmations of the depletion of mtDNA were carried out by PCR analyses on genomic DNA isolated from the 143B.TK−K7 cells with primers that amplify a 1698-bp region located within the D-loop (Figure 3A). Positive controls for the amplification were carried out with genomic DNA of the wild type and as negative control of 143B.TK− ρ0 cells. Additional examinations were accomplished with primers amplifying a region of the nuclear gene histone H1 to proof a sufficient amount of template DNA in the samples. In contrast to the wild-type cell line, no amplification product could be observed in the 143B.TK− ρ0 and 143B.TK−K7 cells. Furthermore, these results were additionally confirmed by Southern blot testing and showed a complete loss of mtDNA in the 143B.TK− ρ0 and 143B.TK−K7 cells (Figure 3B). Low hybridization signals observed as ‘smear’ could be seen in all lanes, suggesting that these may represent hybridization signals to fragments of mtDNA incorporated into the nuclear genome during evolution. Similar results were obtained in multiple independent clones of the LMTK− and NRK52E cell lines (Figure 3C).
Figure 3.
Depletion of mtDNA in different cell lines after expression of a mitochondrially targeted EcoRI. (A) 143B.TK− wild-type and 143B.TK− ρ0 controls as well as the isolated 143B.TK−K7 cells were analysed by PCR using primers corresponding to the D-loop region of mtDNA additionally to primers amplifying a histon H1 of nuclear DNA. It was not possible to amplify this mtDNA region in the cell lines 143B.TK− ρ0 and 143B.TK−K7 confirming the depletion of the mitochondrial genome in these cells. Positive amplification of histone H1 shows a sufficient amount of genomic DNA in all probes. (B) Southern blot analyses with these cells after digestion with BamHI or PvuII using a probe coding for mtDNA nucleotides 4831–5651 also showed the absence of mtDNA in the 143B.TK− ρ0 and 143B.TK−K7 cells (B). (C) LMTK− (mouse) and NRK52E (rat) cells were treated and analysed as described above. mtDNA was undetectable in the newly established ρ0 cell lines of mouse and rat.
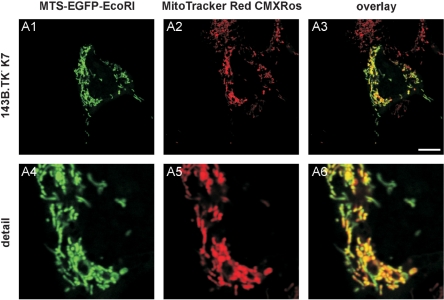
Additional confirmation of the ρ0 state of the cells was acquired by transfecting the ρ0 cell line with the pMEE-con vector. The rationale behind this experiment is that if mtDNA was completely removed, no nucleoid-like structures should appear upon fluorescence microscopic analyses. As shown in Figure 4, the EGFP-tagged restriction enzyme appears as an evenly green-stained mitochondrial network without any punctate pattern. Thus, the punctate appearance of the EcoRI seen originally in the wild-type cells (Figure 1B) accounts for mitochondrial nucleoids, whereas the absence of nucleoids (mtDNA) leads to an even-stained matrix localization of the restriction enzyme, underlining the ρ0 state of the cells.
Figure 4.
143B.TK−K7 cells were transfected with pMEE-con and analysed by confocal fluorescence microscopy. The EGFP-tagged restriction enzyme (MTS-EGFP-EcoRI, green colour, A1 and A4) co-localizes (A3 and A6) with the MitoTracker Red CMXRos stained mitochondrial network (A2 and A5). When compared to Figure 1B, the punctate appearance (‘nucleoid’ structure) of MTS-EGFP-EcoRI merged into an even-stained mitochondrial network, indicating that the interacting partner (mtDNA) of the restriction enzyme (MTS-EGFP-EcoRI) disappeared thus underlining the ρ0 state of the cell.
To analyse whether or not the plasmid was integrated into the host genome during transient transfection, PCR analysis specific for the coding sequence of the restriction endonuclease was carried out on DNA extracts (see Supplementary Data). As expected, no PCR amplification product could be detected (see Figure S1 in Supplementary Data). Thus, an integration of the EcoRI plasmid into the cells nucleus could be excluded.
During more than 1 year of cultivation, there was no recovery of mtDNA in the 143B.TK−K7 cells tested by PCR and metabolic dependence of pyruvate and uridine.
Comparative analyses of the culture medium
The energy production of ρ0 cells completely depends on anaerobic glycolysis. Additionally, all ρ0 cell lines known so far exhibit a dependence on supplementation with pyruvate (3) and uridine (12–14) as well as an increasing acidification of the culture medium by excessive lactic acid production (15). Both characteristic features could be observed in the 143B.TK−K7 cells and the mouse and rat ρ0 clones.
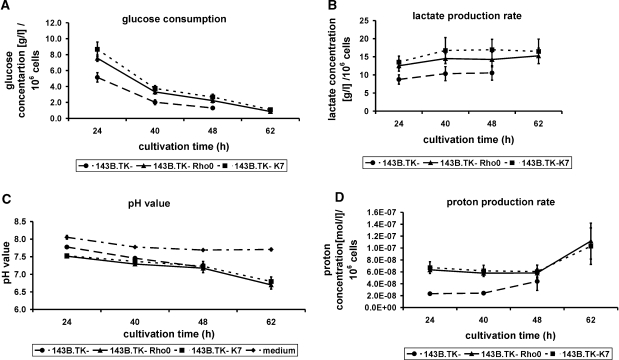
Therefore, the 143B.TK−K7 cells were analysed after 24, 40, 48 and 62 h of cultivation for glucose concentration, lactate concentration and pH value in the culture medium additionally to the determination of cell number. The glucose and lactate concentration as well as the proton concentration were standardized to 106 cells and graphically illustrated. Reference values were obtained by the examination of 143B.TK− and 143B.TK− ρ0 cells.
Standardized to 106 cells, the glucose consumption (Figure 5A) of the ρ0 cells (7.5–8.5 g/l, 143B.TK− ρ0, solid line and 143B.TK−K7, dotted line) exceeds the amount of the wild-type cells (5 g/l, dashed line). Furthermore, it is obvious that the glucose consumption decreases during cultivation time (∼88% in ρ0 cells and 75% in wild-type cells).
Figure 5.
Graphic documentation of the glucose consumption (A), lactate production rate (B) and proton production rate (C) in pure culture medium (dash-dotted line) and the culture medium of the cell lines 143B.TK−K7 (dotted lines) 143B.TK− ρ0 (solid lines) and 143B.TK− (dashed lines) at proceeding cultivation times standardized to 106 cells. The indicated values are averages of four individual measurements, respectively eight independent measurements (pH value of ρ0 cells). The standard deviations are depicted as error bars. Time scale represents the time intervals proceeded after cell seeding. Standardized to 106 cells the glucose consumption as well as the lactate and proton production of the 143B.TK−K7 and 143B.TK− ρ0 cells exceed the amounts of the 143B.TK− wild type. However, the pH value of culture medium in all determined cell lines was nearly similar. The pH value of pure medium without addition of cells shows a slight decline due to saturation of medium with CO2. An especial curve progression could be observed in the analysis of proton production rate where the values initially decline and then increase. It was not possible to determine the 62 h cultivation value of the wild-type cells because of extreme cell density resulting in apoptotic events.
The graphical documentation of lactate production standardized to 106 cells (Figure 5B) shows a higher lactate production rate of the ρ0 cells (143B.TK− ρ0, solid line and 143B.TK−K7, dotted line) than the wild type. While the values initially increase during 24 until 40 h cultivation, the curve progression resembles a saturation thereafter and then stays at an almost constant level (143B.TK−K7: 17 g/l, 143B.TK− ρ0: 15 g/l, 143B.TK−: 11 g/l).
The pH values of pure medium without addition of cells show only a slight decline of pH 8.0 to pH 7.7 due to saturation of medium with CO2 (Figure 5C, dotted-dashed line). On the other hand, the pH value of the medium of the examined cell lines changes from 7.8 in wild-type cells (dashed line) and 7.5 in ρ0 cells (143B.TK− ρ0, solid line and 143B.TK−K7, dotted line) to pH 7.3 after 48 h cultivation time in all cell lines (Figure 5C). Whereas the 62-h value of the 143B.TK− cells could not be determined due to cell mortality that was caused by high cell density, in the cells depleted of mtDNA the pH value further declined to 6.7–6.8.
The proton concentration standardized to the cell number at different cultivation times (Figure 5D) exhibits that the proton-discharging rate in the medium of ρ0 cells (143B.TK− ρ0, solid line and 143B.TK−K7, dotted line) highly exceeds the demand of the wild-type cells (2.5–3.5 × 10−8 mol/l, dashed line). Besides an especial curve progression could be observed in the ρ0 cells where the values initially decline from 7 × 10−8 to 6 × 10−8 mol/l and then increase to 1.0–1.5 × 10−7 mol/l.
Comparative morphological studies with confocal microscopy
The morphology of the mitochondria in the denoted cell lines was studied with confocal microscopy using MitoTracker Red CMXRos to stain the organelles (Figure 6).
Figure 6.
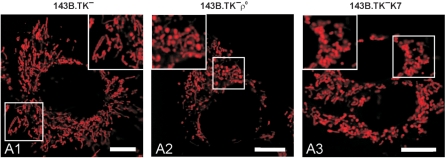
Mitochondrial organization in wild-type and ρ0 143B.TK− cells. The cells were stained with MitoTracker Red CMXRos. (A1) shows the predominantly reticular organization of mitochondria in wild-type cells with mainly rod-shaped organelles. 143B.TK− ρ0 (A2) and 143B.TK−K7 cells (A3) stained with MitoTracker Red CMXRos denotes the disruption of the mitochondrial reticulum in single units that often seemed to be swollen. Calibration marks correspond to 10 µm.
The mitochondria of 143B.TK− wild-type cells (Figure 6A1) exist as reticular network evenly distributed within the cell. The prevalent mitochondrial structure was elongated and rod shaped.
In the ρ0 form of these cells (Figure 6A2 and A3), the network structure appears disrupted, yielding a distribution of small individual mitochondrial units. This fragmentation is highlighted in the higher magnification images. Furthermore, some of these single organelles seemed to be swollen; this result could not be observed in the wild type.
These observations of an interconnected mitochondrial network in wild-type cells in contrast to punctated mitochondria in ρ0 cells constitute typical changes of mitochondrial morphology due to a loss of mitochondrial DNA (16,17).
Comparative morphological studies with transmission electron microscopy (TEM)
Even more morphological changes of the ρ0 mitochondria could be detected by ultra-structural studies (Figure 7).
Figure 7.
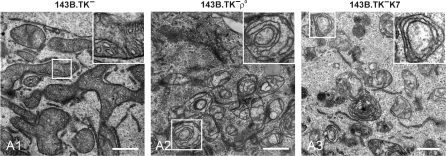
Ultra-structural investigation of wild-type and ρ0 mitochondria by TEM. Electron micrographs of ultra-thin sections are shown. 143B.TK− wild-type mitochondria show an interconnected network structure with numerous regular arranged cristae (A1). The mitochondria of 143B.TK− ρ0 (A2) and 143B.TK−K7 cells (A3) demonstrate single vesicular organelles with distorted cristae, lying in concentric double membrane rings in the matrix. The insets show higher magnifications of the boxed areas. Bars (A1–A3), 1 µm.
Figure 7A1 details a highly interconnected mitochondrial network structure in the wild type. The cross-section through the mitochondrial reticulum in normal 143B.TK− cells showed a distinct outer and inner membrane with electron-dense matrix full of regular arranged cristae structures. In contrast, the TEM images of the ρ0 cells (Figure 7A2 and A3) show an apparent altered morphology of the mitochondria. The network is degraded in single mitochondrial units, often with swollen appearance. The matrix seems to be electron empty probably due to a dilution of the matrix by increasing influx of water that also explains the swollen appearance. The mitochondria in ρ0 cells still exhibit the distinct outer and inner membranes seen in normal cells; but the cristae displayed gross changes of structure. Most cristae appear curved or as concentric rings consisting of two membranes in close contact, an appearance denoted as ‘fuzzy onions’ (18,19). Additionally, these membranous rings showed only few contact sites with the mitochondrial border membrane. It could not be distinguished whether these double membranes consist of two inner membranes or one outer and one inner membrane.
These results also are supported by investigations of other groups (20–25).
DISCUSSION
Very little is known about the key processes of interaction between nucleus and mitochondria, the coordination of gene expression or how mtDNA is maintained within the cell. Resulting from this lack of knowledge, studies were carried out to shed light on the molecular mechanisms underlying mitochondrial dysfunctions. Considerably, work was done to identify nuclear encoded proteins that act on mitochondrial DNA replication and transcription. It is now clear that for example DNA polymerase γ is one of the key players involved in the generation of mtDNA mutations (26–28). Numerous neurological and neuromuscular diseases have been associated with mutations of the mitochondrial genome (1).
In the past, many efforts to overcome these difficulties were carried out by eliminating the mtDNA as one component in the cross talk between the two genomes and thus to generate cells devoid of endogenous mtDNA (so called ρ0 cells). However, all chemicals used so far (ethidium bromide, ditercalinium, ddC, etc.) presented severe disadvantages, such as mutagenic effects on nuclear DNA or the induction of mdr family gene expression (multi-drug resistance). Moreover, the major limitation of these methods is their inapplicability to different cell lines or to create transgenic animal models utilizing this pathway.
The methodical approach of mitochondrially targeted restriction endonucleases for the fragmentation of mitochondrial DNA was pursued in the past by two groups. The successful targeting and function of various restriction endonucleases in the mitochondria of human and murine culture cells could be demonstrated (29,30). Cells with heteroplasmic genotypes were used, where mutated mtDNA harbouring a single base replacement co-exists together with wild-type mitochondrial genomes. The mutated site accounts for a newly formed recognition site for a restriction endonuclease that is not present in the wild-type DNA. Hence, by expressing the appropriate restriction enzyme, the mutated DNA can be selectively fragmented. Upon further degradation of the cleaved molecule by endogenous mitochondrial nucleases, a homoplasmic wild-type genotype can be regained. This method was used with the restriction endonuclease PstI in murine cells harbouring the heteroplasmic mutation for NARP (29,31). Additionally, the restriction endonuclease SmaI was applied, to the mitochondria of human cybrids (30) containing the heteroplasmic recognition sequence associated with Leigh disease (32).
Our strategy to destroy endogenous mtDNA in vivo was also based on a restriction endonuclease that was targeted to the matrix of mitochondria thereby cleaving the genomes and allowing endogenous enzymes to fully disintegrate the DNA molecules so that cells devoid of any endogenous mitochondrial genomes (ρ0 cells) can be generated. This goal was reached by selecting the gene for the restriction endonuclease EcoRI that is known to cleave human mtDNA 3–5 times, depending on ethnic background of the mitochondrial DNA. By fusing the EcoRI gene to a mitochondrial targeting sequence (derived from subunit VIII of human cytochrome c oxidase) and to the EGFP gene (enhanced green fluorescent protein) as optical marker, mitochondrial localization could be visually achieved. Moreover, a selection of transfected cells with FACS analyses can be achieved with the aid of EGFP (data not shown). Utilizing this vector in cell culture, we were able to generate ρ0 cells very efficiently on a 143B.TK− background as well as in mouse and rat cell lines. The loss of all endogenous mitochondrial DNA was monitored both by metabolic (ρ0 cells are kept on regular growth media devoid of traces of uridine and pyruvate) and genetic testing (PCR- or Southern blot analysis). Therefore, it could be shown that this method is applicable not only for the 143B.TK− and LMTK− cell lines that are known to be capable of ρ0 generation via the ethidium bromide method, but also for a rat cell line. The latter result is of interest since there are no published rat ρ0 lines despite anecdotal evidence of attempts by various groups using the ethidium bromide method over the past 15 years. It is possible that our method may be more widely applicable to different cell types and species backgrounds.
After derivation of the ρ0 clone 143B.TK−K7, the EGFP fluorescence of the EcoRI fusion protein could not be detected further. Therefore we tested by PCR if the cells had lost the gene for the restriction endonuclease construct after destruction of the mtDNA or if some gene parts were integrated into the nuclear genome so that continually low amounts of the restriction endonuclease were expressed that would render the cell line useless as acceptor cell line in cytoplast fusion experiments. Analyses with different primer pairs amplifying varying regions of the EcoRI gene showed that no amplification products could be observed. Obviously, the gene for EcoRI was not integrated into the nuclear genome of the 143B.TK−K7 cells. In accordance to the data of Tanaka, our results prove that only a short temporal expression of the restriction endonuclease targeted to the mitochondria is sufficient to destroy completely, irreversibly and stably the mitochondrial genome, also indicating that DNA-repair systems in mitochondria are not efficient enough to cope with the DNA damage.
All generated culture cells depleted of mtDNA so far proliferate despite their lack of oxidative phosphorylation in mitochondria. However, 143B.TK− cells with ρ0 genotype exhibit a decrease in proliferation rate compared to wild type (33). This feature was obvious in the newly established ρ0 cell line 143B.TK−K7 due to the exclusive anaerobic energy production via glycolysis. Thus, all ρ0 cell lines depend on supplementation with pyruvate (3) and uridine (12–14). Additionally, they show an increased acidification of the culture medium by lactic acid (15). Within this work, the dependence on supplementation with pyruvate and uridine was also obvious for the 143B.TK−K7 cells under normal growth conditions as well as an acidification of the culture medium by lactic acid fermentation.
Investigation of the culture medium showed that the pH value of the ρ0 cells was substantially decreased, compared to wild-type medium. While it was not possible to determine the 62 h value of the 143B.TK− cells due to cell mortality that was caused by high cell density, cells depleted of mtDNA exhibited a further decline of the pH to 6.7–6.8.
Standardized to 106 cells, the glucose consumption as well as the lactate and proton production of the 143B.TK−K7 and 143B.TK− ρ0 cells were increased. In contrast to the wild-type cells, this results from a complete dependence of the ρ0 cells on glycolysis for ATP production and therefore an acidification by enhanced lactic acid fermentation occurs. It was shown that in MOLT-4 cells the lactate production was increased 4-fold compared with the wild type (20). However, in the present work the lactic acid production of the cells depleted of mtDNA was only increased 1.5–2-fold in contrast to the wild type.
The transport of lactate and other monocarboxylates within and between cells is accomplished by a family of monocarboxylate transport proteins (MCTs). The mitochondrial lactate/pyruvate exchanger probably collaborates with a mitochondrial lactate dehydrogenase that enables the oxidation of lactate in active respiring cells (34). This pathway cannot take place in ρ0 cells, so that the total amount of lactate produced must be released into the surrounding medium. Across the plasma membrane, lactate is co-transported with protons (35) accounting for the decrease in pH value of the culture medium in the ρ0 cells.
The morphology of the mitochondria in the denoted cell lines was studied with confocal microscopy using MitoTracker Red CMXRos to stain the organelles. The observations of a widespread mitochondrial network in wild-type cells in contrast to punctated mitochondria which often seem to be swollen in ρ0 cells constitute typical changes of mitochondrial morphology due to a loss of mitochondrial genomes (16,17).
Even more morphological changes of the ρ0 mitochondria were detected by ultra-structural studies. In contrast to the mitochondrial reticulum in normal 143B.TK− cells with electron-dense matrix full of regular arranged cristae structures, the TEM images of the ρ0 cells show an apparent altered morphology of the mitochondria. The network is degraded in single mitochondrial units often with swollen appearance and electron empty matrix probably due to more dilution by increasing influx of water. Most cristae exist as ‘fuzzy onions’-like structures (18,36) composed of two membranes lying in concentric rings in the matrix. Additionally, these membranous rings showed only few contact sites with the mitochondrial border membrane. It is not clear whether these double membranes consist of two inner membranes or one outer and one inner membrane.
Similar ρ0 phenotypes with swollen mitochondria with membranous inclusions and multiple concentric cristae could be observed after incubating cells with the reverse transcription inhibitor zidovudine (37). However, by adding of l-carnitine these alterations could be avoided. This indicates a probable involvement of defective fatty acid pathways in these morphological changes.
By applying the new method described here it is possible for the first time to develop new ρ0 cell lines under very controlled and mild conditions, avoiding mutagenic effects of chemicals (e.g. ethidium bromide). Thus, ρ0 cell lines exhibiting varying nuclear backgrounds can be used in future experiments to study diseases connected to mitochondrial phenotypes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
ACKNOWLEDGEMENTS
This work was supported in parts by the Deutsche Forschungsgemeinschaft and the Sächsische Staatsministerium für Wissenschaft und Kunst. The authors are grateful to Paul Modrich and Antal Kiss for the provision of the vector pAN4, Ingrid Rupp, Diana Behrens, Birgit Löffler and Christina ‘Paula’ Paul for technical assistance and to Angela Mühlberg for critical reading of the manuscript. Funding to pay the Open Access publication charges for this article was provided by Genaxxon BioScience GmbH.
Conflict of interest statement. None declared.
REFERENCES
- 1.Wallace DC. A mitochondrial paradigm for degenerative diseases and ageing. Novartis Found. Symp. 2001;235:247–263. doi: 10.1002/0470868694.ch20. [DOI] [PubMed] [Google Scholar]
- 2.Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 3.King MP, Attardi G. Human cells lacking mtDNA: repopulation with exogenous mitochondria by complementation. Science. 1989;246:500–503. doi: 10.1126/science.2814477. [DOI] [PubMed] [Google Scholar]
- 4.Borst P, Grivell LA. The mitochondrial genome of yeast. Cell. 1978;15:705–723. doi: 10.1016/0092-8674(78)90257-x. [DOI] [PubMed] [Google Scholar]
- 5.Goldring ES, Grossman LI, Krupnick D, Cryer DR, Marmur J. The petite mutation in yeast. Loss of mitochondrial deoxyribonucleic acid during induction of petites with ethidium bromide. J. Mol. Biol. 1970;52:323–335. doi: 10.1016/0022-2836(70)90033-1. [DOI] [PubMed] [Google Scholar]
- 6.Nagley P, Linnane AW. Mitochondrial DNA deficient petite mutants of yeast. Biochem. Biophys. Res. Commun. 1970;39:989–996. doi: 10.1016/0006-291x(70)90422-5. [DOI] [PubMed] [Google Scholar]
- 7.Inoue K, Ito S, Takai D, Soejima A, Shisa H, LePecq JB, Segal-Bendirdjian E, Kagawa Y, Hayashi JI. Isolation of mitochondrial DNA-less mouse cell lines and their application for trapping mouse synaptosomal mitochondrial DNA with deletion mutations. J. Biol. Chem. 1997;272:15510–15515. doi: 10.1074/jbc.272.24.15510. [DOI] [PubMed] [Google Scholar]
- 8.Inoue K, Takai D, Hosaka H, Ito S, Shitara H, Isobe K, LePecq JB, Segal-Bendirdjian E, Hayashi J. Isolation and characterization of mitochondrial DNA-less lines from various mammalian cell lines by application of an anticancer drug, ditercalinium. Biochem. Biophys. Res. Commun. 1997;239:257–260. doi: 10.1006/bbrc.1997.7446. [DOI] [PubMed] [Google Scholar]
- 9.Meyer RR, Simpson MV. DNA biosynthesis in mitochondria. Differential inhibition of mitochondrial and nuclear DNA polymerases by the mutagenic dyes ethidium bromide and acriflavin. Biochem. Biophys. Res. Commun. 1969;34:238–244. doi: 10.1016/0006-291x(69)90637-8. [DOI] [PubMed] [Google Scholar]
- 10.Nelson I, Hanna MG, Wood NW, Harding AE. Depletion of mitochondrial DNA by ddC in untransformed human cell lines. Somat. Cell Mol. Genet. 1997;23:287–290. doi: 10.1007/BF02674419. [DOI] [PubMed] [Google Scholar]
- 11.Newman AK, Rubin RA, Kim SH, Modrich P. DNA sequences of structural genes for Eco RI DNA restriction and modification enzymes. J. Biol. Chem. 1981;256:2131–2139. [PubMed] [Google Scholar]
- 12.Gregoire M, Morais R, Quilliam MA, Gravel D. On auxotrophy for pyrimidines of respiration-deficient chick embryo cells. Eur. J. Biochem. 1984;142:49–55. doi: 10.1111/j.1432-1033.1984.tb08249.x. [DOI] [PubMed] [Google Scholar]
- 13.Morais R, Giguere L. On the adaptation of cultured chick embryo cells to growth in the presence of chloramphenicol. J. Cell Physiol. 1979;101:77–88. doi: 10.1002/jcp.1041010110. [DOI] [PubMed] [Google Scholar]
- 14.Morais R, Gregoire M, Jeannotte L, Gravel D. Chick embryo cells rendered respiration-deficient by chloramphenicol and ethidium bromide are auxotrophic for pyrimidines. Biochem. Biophys. Res. Commun. 1980;94:71–77. doi: 10.1016/s0006-291x(80)80189-6. [DOI] [PubMed] [Google Scholar]
- 15.Jakobs BS, van den BC, Dacremont G, Wanders RJ. Beta-oxidation of fatty acids in cultured human skin fibroblasts devoid of the capacity for oxidative phosphorylation. Biochim. Biophys. Acta. 1994;1211:37–43. doi: 10.1016/0005-2760(94)90136-8. [DOI] [PubMed] [Google Scholar]
- 16.Gilkerson RW, Margineantu DH, Capaldi RA, Selker JM. Mitochondrial DNA depletion causes morphological changes in the mitochondrial reticulum of cultured human cells. FEBS Lett. 2000;474:1–4. doi: 10.1016/s0014-5793(00)01527-1. [DOI] [PubMed] [Google Scholar]
- 17.Rizzuto R, Pinton P, Carrington W, Fay FS, Fogarty KE, Lifshitz LM, Tuft RA, Pozzan T. Close contacts with the endoplasmic reticulum as determinants of mitochondrial Ca2+ responses. Science. 1998;280:1763–1766. doi: 10.1126/science.280.5370.1763. [DOI] [PubMed] [Google Scholar]
- 18.Hales KG, Fuller MT. Developmentally regulated mitochondrial fusion mediated by a conserved, novel, predicted GTPase. Cell. 1997;90:121–129. doi: 10.1016/s0092-8674(00)80319-0. [DOI] [PubMed] [Google Scholar]
- 19.Halestrap AP. The regulation of the matrix volume of mammalian mitochondria in vivo and in vitro and its role in the control of mitochondrial metabolism. Biochim. Biophys. Acta. 1989;973:355–382. doi: 10.1016/s0005-2728(89)80378-0. [DOI] [PubMed] [Google Scholar]
- 20.Armand R, Channon JY, Kintner J, White KA, Miselis KA, Perez RP, Lewis LD. The effects of ethidium bromide induced loss of mitochondrial DNA on mitochondrial phenotype and ultrastructure in a human leukemia T-cell line (MOLT-4 cells) Toxicol. Appl. Pharmacol. 2004;196:68–79. doi: 10.1016/j.taap.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 21.Herzberg NH, Middelkoop E, Adorf M, Dekker HL, Van Galen MJ, Van den BM, Bolhuis PA, van den BC. Mitochondria in cultured human muscle cells depleted of mitochondrial DNA. Eur. J. Cell Biol. 1993;61:400–408. [PubMed] [Google Scholar]
- 22.Holmuhamedov E, Jahangir A, Bienengraeber M, Lewis LD, Terzic A. Deletion of mtDNA disrupts mitochondrial function and structure, but not biogenesis. Mitochondrion. 2003;3:13–19. doi: 10.1016/S1567-7249(03)00053-9. [DOI] [PubMed] [Google Scholar]
- 23.Morais R, Desjardins P, Turmel C, Zinkewich-Peotti K. Development and characterization of continuous avian cell lines depleted of mitochondrial DNA. In Vitro Cell Dev. Biol. 1988;24:649–658. doi: 10.1007/BF02623602. [DOI] [PubMed] [Google Scholar]
- 24.Porteous WK, James AM, Sheard PW, Porteous CM, Packer MA, Hyslop SJ, Melton JV, Pang CY, Wei YH, et al. Bioenergetic consequences of accumulating the common 4977-bp mitochondrial DNA deletion. Eur. J. Biochem. 1998;257:192–201. doi: 10.1046/j.1432-1327.1998.2570192.x. [DOI] [PubMed] [Google Scholar]
- 25.van den Ouweland JM, Maechler P, Wollheim CB, Attardi G, Maassen JA. Functional and morphological abnormalities of mitochondria harbouring the tRNA (Leu)(UUR) mutation in mitochondrial DNA derived from patients with maternally inherited diabetes and deafness (MIDD) and progressive kidney disease. Diabetologia. 1999;42:485–492. doi: 10.1007/s001250051183. [DOI] [PubMed] [Google Scholar]
- 26.Falkenberg M, Larsson NG, Gustafsson CM. DNA replication and transcription in mammalian mitochondria. Annu. Rev. Biochem. 2007;76:679–699. doi: 10.1146/annurev.biochem.76.060305.152028. [DOI] [PubMed] [Google Scholar]
- 27.Hudson G, Chinnery PF. Mitochondrial DNA polymerase-gamma and human disease. Hum. Mol. Genet. 2006;15:R244–R252. doi: 10.1093/hmg/ddl233. [DOI] [PubMed] [Google Scholar]
- 28.Copeland WC. Inherited mitochondrial diseases of DNA replication. Annu. Rev. Med. 2008;59:131–146. doi: 10.1146/annurev.med.59.053006.104646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Srivastava S, Moraes CT. Manipulating mitochondrial DNA heteroplasmy by a mitochondrially targeted restriction endonuclease. Hum. Mol. Genet. 2001;10:3093–3099. doi: 10.1093/hmg/10.26.3093. [DOI] [PubMed] [Google Scholar]
- 30.Tanaka M, Borgeld HJ, Zhang J, Muramatsu S, Gong JS, Yoneda M, Maruyama W, Naoi M, Ibi T, et al. Gene therapy for mitochondrial disease by delivering restriction endonuclease SmaI into mitochondria. J. Biomed. Sci. 2002;9:534–541. doi: 10.1159/000064726. [DOI] [PubMed] [Google Scholar]
- 31.Holt IJ, Harding AE, Morgan-Hughes JA. Deletions of muscle mitochondrial DNA in patients with mitochondrial myopathies. Nature. 1988;331:717–719. doi: 10.1038/331717a0. [DOI] [PubMed] [Google Scholar]
- 32.DeVivo DC, Haymond MW, Obert KA, Nelson JS, Pagliara AS. Defective activation of the pyruvate dehydrogenase complex in subacute necrotizing encephalomyelopathy (Leigh disease) Ann. Neurol. 1979;6:483–494. doi: 10.1002/ana.410060605. [DOI] [PubMed] [Google Scholar]
- 33.King MP, Attardi G. Isolation of human cell lines lacking mitochondrial DNA. Methods Enzymol. 1996;264:304–313. doi: 10.1016/s0076-6879(96)64029-4. [DOI] [PubMed] [Google Scholar]
- 34.Brooks GA. Lactate shuttles in nature. Biochem. Soc. Trans. 2002;30:258–264. doi: 10.1042/. [DOI] [PubMed] [Google Scholar]
- 35.Poole RC, Halestrap AP. Transport of lactate and other monocarboxylates across mammalian plasma membranes. Am. J. Physiol. 1993;264:C761–C782. doi: 10.1152/ajpcell.1993.264.4.C761. [DOI] [PubMed] [Google Scholar]
- 36.Hermann GJ, Thatcher JW, Mills JP, Hales KG, Fuller MT, Nunnari J, Shaw JM. Mitochondrial fusion in yeast requires the transmembrane GTPase Fzo1p. J. Cell Biol. 1998;19:359–373. doi: 10.1083/jcb.143.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Semino-Mora MC, Leon-Monzon ME, Dalakas MC. The effect of L-carnitine on the AZT-induced destruction of human myotubes. Part II: treatment with L-carnitine improves the AZT-induced changes and prevents further destruction. Lab. Invest. 1994;71:773–781. [PubMed] [Google Scholar]