Abstract
The quest for inducers and inhibitors of protein amyloidogenesis is of utmost interest, since they are key tools to understand the molecular bases of proteinopathies such as Alzheimer, Parkinson, Huntington and Creutzfeldt–Jakob diseases. It is also expected that such molecules could lead to valid therapeutic agents. In common with the mammalian prion protein (PrP), the N-terminal Winged-Helix (WH1) domain of the pPS10 plasmid replication protein (RepA) assembles in vitro into a variety of amyloid nanostructures upon binding to different specific dsDNA sequences. Here we show that di- (S2) and tetra-sulphonated (S4) derivatives of indigo stain dock at the DNA recognition interface in the RepA-WH1 dimer. They compete binding of RepA to its natural target dsDNA repeats, found at the repA operator and at the origin of replication of the plasmid. Calorimetry points to the existence of a major site, with micromolar affinity, for S4-indigo in RepA-WH1 dimers. As revealed by electron microscopy, in the presence of inducer dsDNA, both S2/S4 stains inhibit the assembly of RepA-WH1 into fibres. These results validate the concept that DNA can promote protein assembly into amyloids and reveal that the binding sites of effector molecules can be targeted to inhibit amyloidogenesis.
INTRODUCTION
Aggregation of proteins into amyloid assemblies is the conundrum of an increasing number of proteinopathies with devastating impact on human health. Although amyloids share related 3D structures, the factors responsible for inducing protein amyloidogenesis are diverse (1). Among the latter, nucleic acid ligands have been extensively studied for the mammalian prion protein (PrP), the causative agent of lethal transmissible spongiform amyloid encephalopathies (2). The conformation of the cellular form of this protein (PrPc) is transformed into its pathogenic variant (PrPsc) upon binding to long, mixed DNA or RNA sequences in vitro, promoting its assembly into amyloid fibres and spheroids (3–7). More recently, the interaction of PrP with a short, specific dsDNA sequence has been characterized by NMR spectroscopy (8) and the in vitro selection of small ssDNA thioaptamers binding to PrP reported (9). The latter study points to the existence in PrP of two DNA-binding sites, one with high affinity (specific) and another with lower affinity (non-specific) (9). The reports by Supattapone and co-workers (10), on the strict requirement of polyanionic additives for the successful amplification in vitro of PrPsc from recombinant PrP and on the existence of a stable complex constituted by several PrP molecules bound to RNA/ssDNA (11), have the highest relevance for understanding prion amyloidogenesis.
RepA is a multifunctional DNA binding protein encoded by the Pseudomonas pPS10 plasmid (12). RepA dimers bind to repA gene operator repressing its own transcription, whereas RepA monomers activate plasmid replication after cooperative binding to four directly repeated DNA sequences (termed iterons) (13). Single iteron binding, by itself, enhances dissociation of RepA dimers and induces a structural transformation (14,15) in the N-terminal Winged-Helix dimerization domain (WH1) (16) that implies the conversion of part of its α-helical elements into β-strands and loops (17). Although stable binding of RepA to dsDNA requires the presence of a second, C-terminal WH domain (WH2), both WH1 and WH2, when isolated, can bind to their targets at the operator and iteron sequences (16). The conformational changes experienced by RepA, paralleling those in known amyloid forming proteins (1), have recently inspired a search for conditions leading to RepA-WH1 amyloidogenesis in vitro (18). Specific operator or iteron core sequences (11 bp), combined with point mutations in an amyloidogenic sequence located opposite to the DNA-binding interface (17), drive the assembly of the domain into a range of amyloid nano-structures spanning from irregular aggregates to well ordered fibres, through regular spheroids (18). The key determinant for the final type of assembly or ‘strain’ is the sequence of the dsDNA ligand (operator for the fibres), albeit DNA itself is not a component of the matured amyloids (18). This suggests that transient binding of a specific dsDNA to WH1 would shield an electropositive patch at the DNA binding interface, thus assisting the ordered assembly of the protein into fibres. The latter is achieved through a cross-β spine made of the amyloidogenic peptide sequence L26VLCAVSLI34, distal to the dsDNA-binding site (18).
RepA-WH1 provides a suitable and novel bacterial model system to study sequence-specific DNA-promoted protein amyloidogenesis. Here, we have searched for small molecules docking in silico to RepA-WH1 and found di- (S2) and tetra-sulphonated (S4) derivatives of the classic indigo stain that compete with RepA binding to dsDNA. The thermodynamic characterization of S4-indigo interaction shows that it involves a major binding site in WH1 (Kd = 0.23 μM). Inhibition by the sulphonated indigoids of RepA-WH1 binding to the operator dsDNA sequence abolishes fibre assembly, constituting a proof of concept for DNA-induced protein amyloidogenesis (18). Unlike the wealth of small molecules described so far to inhibit protein aggregation into amyloids (19,20), those reported here are the first acting at the binding site for a regulator or enhancer of amyloidogenesis, rather than at the peptide sequences directly assembling into the landmark amyloid cross-β structure.
MATERIALS AND METHODS
Docking of sulphonated indigoids to RepA-WH1
The chemical structures of indigo (3,3′-dioxo-2,2′-bis-indole) and its 5,5′-disulphonic (indigotine or indigo carmine) and 5,5′,7,7′-tetrasulphonic derivatives were outlined by using ChemDraw (v.10.0, CambridgeSoft) and then exported to SMILES code. The 3D structures of the molecules were generated by means of Corina (http://www.molecular-networks.com/online_demos/corina_demo.html) and saved as PDB files. Docking of the indigoid molecules to the structure of RepA-WH1 dimer (17; PDB code 1HKQ) was performed using AutoDock (v.3.05) (21), run in an Apple workstation (2 × 2 GHz PowerPC G5, OSX 10.3.9). Being the indigo (bis-indolic) core planar considerably simplifies the flexible docking of the ligands, because it reduces the number of torsional degrees of freedom that have to be taken into account to those concerning the four (S2-indigo) or eight (S4-indigo) substituent sulphate groups. Protein structure was screened within a 50 × 50 × 50 Å cube centred at the dimer interface and sampled with a grid in which points were spaced 0.425 Å. Translation and rotation steps were set to 1.5 Å and 25°, respectively. A Lamarckian genetic algorithm (mutation rate 0.02, crossover rate 0.8) was then used for 25 runs in a population of 50 individuals. All the solutions fitted to the DNA binding interface in RepA-WH1 (17). Independent runs placing the sampled volume around other protein areas did not yield a solution with a noticeable low ΔG.
Protein purification and DNA synthesis
RepA-WT protein and its isolated WH1 domain, either WT or carrying the amyloidogenic enhancing mutation A31V, were purified as described (16,18). Concentrated protein stocks were dialysed against 0.1 M Na2SO4, 20 mM Tris–SO4 pH 8, 4 mM MgSO4, 1 mM 2-MeEtOH, 10% glycerol before long-term storage at –70°C. Protein concentration, determined by A280 nm (14,16), is expressed in a per-monomer basis. The dsDNA oligonucleotide opsp (5′-CATTGACTTGT/3′-GTAACTGAACA) (18) was synthesized and annealed (0.2 mM stock) as previously described (16). For the DNA binding competition assays by indigoids, PCR was performed using as templates pUC18 derivatives (0.1 μg) carrying either a 39 bp sequence spanning the repA operator or a 22 bp single pPS10-oriV iteron, cloned into the vector SmaI site (14,15). The universal primers f17 and r19 (50 pmol each) were used in 40 cycles of amplification by gel-adsorbed Taq DNApol (BioTools). Amplified dsDNAs were purified by means of phenol–chloroform–isoamylic alcohol extraction plus ethanol precipitation.
Electrophoretic mobility shift assays (EMSA)
Ten microlitre aliquots containing 35 pmol of RepA-WT in protein buffer (see above) were incubated for 15 min on ice with serial dilutions of S2/S4-indigo (purchased from TCI Europe, Belgium; 10 mM stocks in water). Then the operator or iteron dsDNAs (calibrated to be ≈50% bound by RepA) were added and incubated for further 15 min on ice, before adding 2.5 μl of 0.05% bromophenol blue in 20% glycerol. Samples were immediately loaded onto 6% polyacrylamide (29:1)–0.25 × TBE minigels. Electrophoresis was run at 150 V, for 1 h at 4°C. After ethidium bromide staining, bands were quantified by UV detection (Bio-Rad Gel Doc 2000).
Isothermal titration calorimetry (ITC)
Six microlitre aliquots of a RepA-WH1-WT stock (150 μM) were extensively dialysed (5 l total volume, various changes for 48 h) against 20 mM Tris–HCl pH 8 at 4°C. A quantity of 1.8 ml of the protein solution (a concentration range spanning 75–100 μM was tested in several independent assays) were displayed into a MicroCal VP-ITC (22), degassed for 5–10 min and equilibrated at 16°C. The reference cell contained buffer from the last dialysis passage. Twenty-five injections (10 μl each) of distinct concentrations (0.62–1.41 mM, diluted in dialysis buffer from a 10 mM stock) of S2/S4-indigo were performed using the control software of the calorimeter, leaving 150 s between injections to allow for equilibration. Baselines were acquired (the buffer at the sample cell titrated with S2/S4-indigo) and subtracted before integration of data. Calculation of the affinity constants (Ka), enthalpy (ΔH) and stoichiometry (n) were performed, according to pre-established binding models, by means of non-linear regression best fitting (23; equation 6b therein), as implemented in the Origin-7 software (Microcal).
Transmission electron microscopy (TEM)
RepA-WH1-A31V (35 μM) fibres were assembled in the presence of equimolar amounts of opsp-dsDNA as described (18), with the only modification of including 1-, 3- or 9-fold molar excess of S2/S4-indigo over protein molecules in the incubations (4°C for 1 month). TEM was performed after negative staining (2% uranyl acetate) using a JEOL JEM-1230 electron microscope, operating at 100 kV (18).
RESULTS
Small molecules targeting the dsDNA binding surface in RepA-WH1
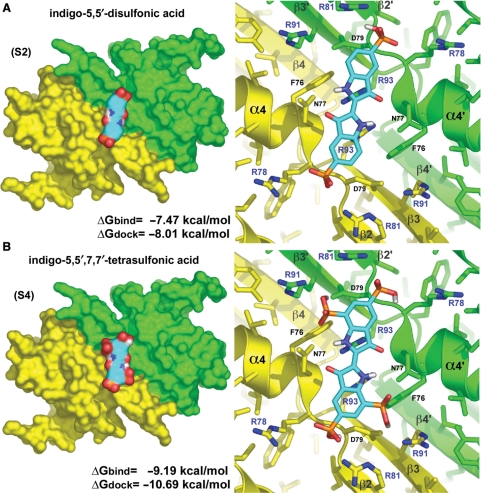
The rationale for the selection of molecules inhibiting RepA-WH1 interaction with dsDNA and thus protein amyloidogenesis (18) was the following. Contacts of WH1 with the phosphate backbone of dsDNA are required for the stable binding of RepA to the operator and iteron sequences (16,24). Thus, the electropositive patch defined in the 3D structure of WH1 dimers (17) by the side chains of arginine residues 78, 81, 91 and 93 (Figure 1) would provide a pocket where an acidic ligand should potentially bind. Since such Arg residues cluster around the pseudo-2-fold symmetry axis in the WH1 dimer (17), the candidate ligand should also exhibit that symmetry. Therefore, a search for small, symmetric, acidic molecules was carried out by visual inspection of the available commercial catalogues. Up to 10 molecules following these criteria, most of them di-carboxylates derived from tartaric acid but also including two sulphonated variants of the blue stain indigo, were tested in silico by docking to the crystal structure of RepA-WH1 dimer (see Materials and Methods section). Those with the best (lowest) ΔG values predicted were the S2 and S4-indigoids (Figure 1), in particular the latter (Figure 1B). The enthalpic term, provided by salt bridges between the electronegative oxygens in the ligand sulphates and the electropositive guanidinium groups in the protein Arg residues, is expected to drive binding, thus tuning the specificity of the interaction. The rigidity of the ligand core leaves the overall entropic term of interaction to rely on the water molecules/ions displaced upon binding.
Figure 1.
Sulphonated indigo derivatives target the dsDNA binding interface in RepA-WH1. Left: the most symmetrically arranged docking solutions for the acidic indigo derivatives (cyan) S2 (A) and S4 (B) on the pseudo-2-fold axis of the protein dimer (17; PDB entry 1HKQ; subunits in yellow and green). The calculated values of ΔG for docking and binding are displayed. Right: close-up views of the sulphonated indigos in electrostatic interaction with the side chains of the Arg residues 78, 81 and 91. The guanidinium group of each Arg93 residue could make a cation-π interaction with its nearest indol moiety that, together with H-bonding between the side chain of Asn77 and the pyrrolidin keto and amino groups, would account for the ΔGdock predicted for the unmodified indigo molecule (−5.54 kcal/mol, data not shown). Models were rendered using PyMOL (http://www.pymol.org).
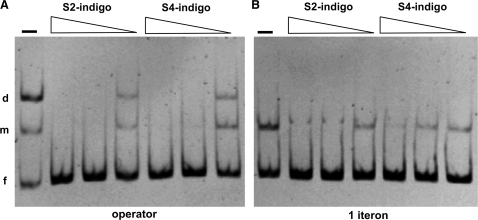
The S2/S4-indigoids were then purchased and their abilities to inhibit the binding of full-length RepA-WT dimers to operator (Figure 2A) and a single iteron (Figure 2B) dsDNA sequences (14–16) were tested in vitro by means of electrophoretic mobility shift assays (EMSA). Both indigoids completed the formation of RepA-operator complexes, with an apparent IC50 between 50–100 μM. In contrast, the tartrate ligand derivatives mentioned earlier worked in a much poorer way (IC50 approaching to millimolar, data not shown). Being the electric field applied in EMSA disruptive for Coulombic interactions in protein–ligand complexes, in particular until these enter into the gel (15), the reported IC50 values are likely overestimated. Besides this, we have not observed any major difference in IC50 if the dyes were added to the reaction mixture after pre-incubating protein with dsDNA, suggesting Kd values for RepA-indigoids binding in the order of those for the specific DNAs (16). It is noteworthy that inhibition by both indigoids of RepA binding to the iteron sequence seems to be less efficient than to the operator inverted repeat. As shown in Figure 1, S2/S4-indigos are expected to bind to RepA dimers (the species binding to the operator) but not to RepA monomers (active in iteron binding), thus the dyes would have on the latter only an indirect effect, consisting in shielding a fraction of the dimers from iteron-induced dissociation into monomers (14,15). In addition, S2-indigo has a net tendency to induce RepA aggregation (part of the dye does not enter into the gel): this could have some contribution to its competitor effect on protein binding to either dsDNA sequence, thus appearing to match the more specific inhibitor S4-indigo.
Figure 2.
EMSA showing competition by S2/S4-indigos (1, 0.25 and 0.063 mM) of the binding of RepA-WT to either a DNA fragment containing the repA operator sequence (A) or a single iteron origin repeat (B). The positions of the free dsDNAs (f) and their complexes with RepA monomers (m) and dimers (d) are indicated.
Thermodynamics of S2/S4-indigos binding to RepA-WH1
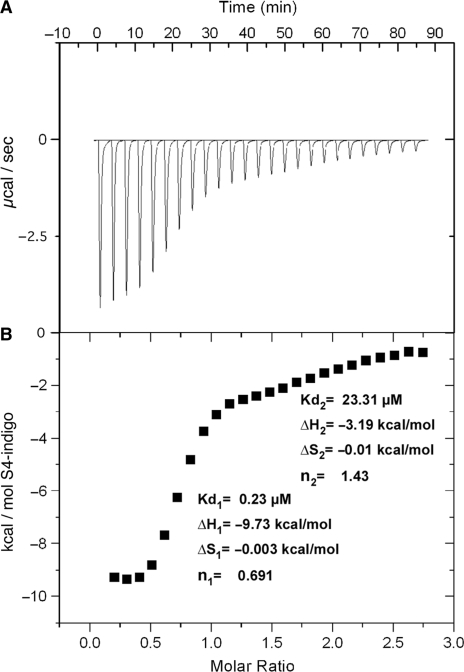
Isothermal Titration Calorimetry (ITC; 23) was then used to get a reliable measure of the binding constants and overall energetics of the interactions between S2/S4 indigoids and RepA-WH1 dimers. The latter was used, rather than full length RepA, due to its higher solubility (16). The titrations performed with S4-indigo (Figure 3) were exothermic, pointing to a major site (Kd = 1/Ka =0.23 μM) with a single molecule bound per protein dimer (ligand-to-protein monomer stoichiometry, n = 0.69) (23), as predicted by docking (Figure 1B). This high affinity protein–ligand interaction is enthalpy-driven (ΔH =−9.73 kcal/mol; ΔS = −0.003 kcal/mol). The free energy of binding calculated from the ITC measurements (ΔG = ΔH–TΔS = −8.86 kcal/mol) is close to that estimated in the docking procedure (ΔG = −9.19 kcal/mol) (Figure 1B), supporting the reliability of the latter. A second transition became evident at the highest concentration range of S4-indigo tested, probably reflecting multiple and non-specific ligand binding (Kd =23.3 μM, n = 1.43; approximately three molecules bound per WH1 dimer; ΔG = 0.3 kcal/mol), which results in the formation of aggregates. We cannot exclude that these non-specific binding sites in RepA-WH1 could become available due to some structural change in the protein induced by the primary S4-indigo binding event. Titration of WH1 with S2-indigo did not allow a reliable determination of either affinity constants or stoichiometry, due to non-specific binding which also gave way to protein aggregation.
Figure 3.
Thermodynamic characterization (ITC) of the binding of S4-indigo to WH1-WT. (A) Binding isotherm for the titration of protein (98 μM) with the ligand (1.41 mM reservoir; concentration interval: 7.8–172 μM). (B) Integration of data plotted against the molar ratio between S4-ligand and WH1 (as monomers). The curve fits to a major binding site (#1) with a secondary site (#2) becoming evident at higher dye:protein ratios.
S2/S4-indigos inhibit WH1-A31V assembly into amyloids
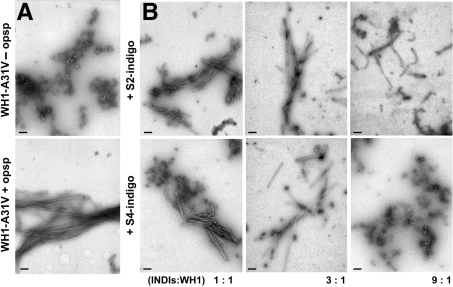
The ability of the S2/S4-indigo molecules to inhibit the dsDNA-induced assembly of the hyper-amyloidogenic WH1-A31V mutant into fibres was then checked. In summary, purified protein and dsDNA oligonucleotide were incubated in the presence of equimolar amounts, or 3- or 9-fold molar excess, of the indigoid ligands for a month, to allow for full maturation of the fibres (18). Samples were then inspected by means of transmission electron microscopy (TEM) after negative staining (Figure 4). In the control samples incubated in the absence of either of the indigoid stains, bundles of long, 75 nm thick, amyloid fibres (18), were evident (Figure 4A). However, samples incubated with S2-indigo (up to 3:1 ligand-to-protein ratio) showed smaller and thinner (ø = 25 nm) fibres. These became even shorter and curved at a higher molar ratio (9:1). When S4-indigo was the molecule tested, the result was very similar to that for the di-sulphonated molecule at the low molar ratios (Figure 4B), but at the highest (9:1) no fibre was evident, rendering instead an amorphous aggregate, similar to that reported (18) when WH1-A31V is incubated in the absence of dsDNA (Figure 4A). In conclusion, targeting with acidic indigo derivatives the binding site of effector dsDNA molecules in WH1-A31V is an efficient way to inhibit the assembly of the latter into amyloid fibres.
Figure 4.
S2/S4-indigos inhibit DNA-promoted WH1-A31V amyloidogenesis according to TEM (magnification bars: 200 nm). (A) Control aggregates (top) and fibres (bottom) observed after 1 month incubation in the absence or presence of opsp dsDNA, respectively (18). (B) Samples including three different indigoid-to-protein (and dsDNA) molar ratios. S2 (top row) and, specially, S4-indigo (bottom row) lead to shorter and thinner fibres and, at the highest concentration tested, S4-indigo yields protein aggregates alike those found in the absence of DNA (see A).
DISCUSSION
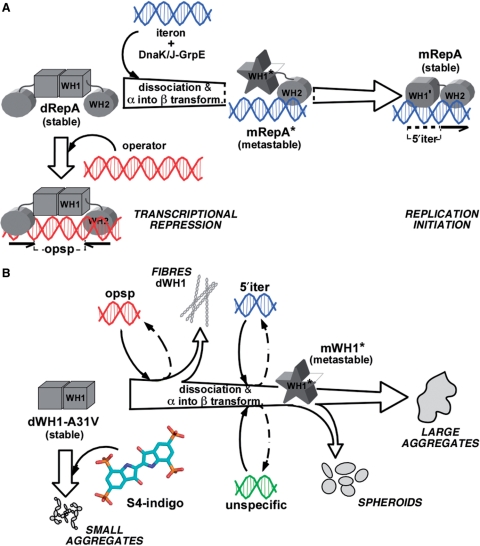
A central issue in current protein science is the survey of factors promoting amyloidogenesis. Natural and synthetic polyanions, in particular RNA and DNA, have been found to enhance the assembly of the mammalian PrP into amyloids (3–11). Similarly, the dimeric WH1 domain of the bacterial plasmid replication protein RepA, besides its role in regulating replication initiation by the full length protein (13; Figure 5A), assembles into distinct amyloid aggregates, including spherulites and ordered fibres, upon transient binding to small specific dsDNA sequences (18) (Figure 5B). These are not constituents of the final aggregated particles which are mainly (if not exclusively) formed by RepA-WH1 (18), in agreement with the protein only nature of any amyloid structure (1,2), assembled through an amyloidogenic motif located opposite to the DNA-binding surface in WH1 structure (17).
Figure 5.
A scheme of the conformational transactions experienced by pPS10 RepA protein. (A) For the whole RepA protein, the balance between dimers (dRepA) and monomers (mRepA) is modulated by the target iteron DNA sequence (blue). WH2 binding implies dimer dissociation and a local secondary structure conversion (α → β) in WH1 (13,16), which becomes a metastable intermediate (the unbound domain depicted as a star) (14). This short-lived species is stabilized through WH1 binding to the iteron (15). On the contrary, dRepA can bind to the operator DNA sequence (red) without altering its association state and with minor effects (if any) on the structure of the WH1 domain (represented as a cube). Overall, the binding to the iteron sequence defines an irreversible refolding trajectory for WH1. (B) The extent of the remodelling exerted by the core of those target DNA sequences in the WH1 domain, in combination with the amyloidogenic enhancer A31V mutation, lead to the assembly of the protein into either ordered fibres, or spheroids, or large and irregular aggregates (18). Their formation can be interpreted as paralleling the pathway outlined in (A). As it is shown in this article, binding of sulphonated indigoids (S4 molecule depicted) to WH1 dimers competes with dsDNA binding, thus inhibiting amyloidogenesis and leading to small, grainy protein aggregates.
We have found that di- and tetra-sulphonated derivatives of the indigo stain dock at the dsDNA recognition surface in RepA-WH1 (17), thus competing protein–DNA interactions and subsequently inhibiting the assembly of WH1 into amyloid fibres. The interaction of a single S4-indigo molecule with a WH1 dimer primarily occurs through a binding site exhibiting micromolar affinity. It can be competed by increasing the ionic strength (becoming undetectable by ITC at 0.1 M Na2SO4; data not shown), as expected from the predicted major enthalpic contribution of electrostatic interactions to ΔG. However, the sulphonated indigoids can still exert their inhibitory action on WH1 amyloidogenesis during the prolonged incubations required for nucleation and growth of the protein fibres. The interaction of S4-indigo with WH1 dimers is specific, albeit at high ligand concentrations multiple molecules bind non-specifically. This would be related to the kind of large micellar complexes described for some hits coming out frequently in high throughput screenings of diverse protein targets (25).
Our results suggest that the promoting action of dsDNA on WH1 amyloidogenesis does not merely consist in shielding the electropositive patch in the protein (17) to allow for fibre assembly (18), since the indigoids would have the same effect and yet they do not yield ordered fibres, but increasingly disordered structures and, finally, amorphous aggregates. Thus, transient interactions with operator dsDNA should promote in WH1 some structural alteration that must be crucial for amyloidogenesis, contributing to the changes in the CD spectra previously attributed just to the assembly of the amyloidogenic stretch into a cross-β spine (18).
Many small molecules, including a number of indol derivatives (26), have been described to directly interfere with protein aggregation through amyloidogenic stretches (19,20). However, to our knowledge those reported here are the first targeting a protein region not directly assembled into a cross-β structure, but a site recognized by an allosteric effector of protein amyloidogenesis. The sulphonated indigoids, due to their specificity for the dsDNA binding pocket in RepA-WH1, are unlikely to have general applicability for other amyloidogenic proteins or even to bind to close RepA homologues, such as the dimers of the RepE protein, which do not bind to operator DNA through the WH1 domain (27). This is probably due to its reduced electropositive character, derived from the lack of basic residues equivalent to Arg81, 91 and 93 in RepA. The S2/S4-indigos also have the intrinsic limitations to membrane permeability derived from their acidic character. Notwithstanding, this work illustrates both the contribution of co-factors (dsDNA) to the assembly of protein amyloids and the feasibility of targeting their binding sites to design anti-amyloidogenic drugs.
ACKNOWLEDGEMENTS
We are indebted to the staff of the oligonucleotide synthesis and electron microscopy facilities at CIB for their technical support, to Dr Javier Ruiz for suggestions on ITC and to Drs Federico Gago and Antonio Romero for encouragement. Our groups are part of the Bioinformatics Integrative Platform for structurE-based Drug Discovery (BIPEDD-CM), funded by the Regional Government of Madrid (P-BIO-0214-2006). This work has been financed by grants of Spanish MEC (BMC2003-00088 and BFU2006-00494) to R.G. Funding to pay the Open Access publication charges for this article was provided by grant BFU2006-00494.
Conflict of interest statement. None declared.
REFERENCES
- 1.Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 2.Prusiner SB. Prions. Proc. Natl Acad. Sci. USA. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cordeiro Y, Machado F, Juliano L, Juliano MA, Brentani RR, Foguel D, Silva JL. DNA converts cellular prion protein into the β-sheet conformation and inhibits prion peptide aggregation. J. Biol. Chem. 2001;276:49400–49409. doi: 10.1074/jbc.M106707200. [DOI] [PubMed] [Google Scholar]
- 4.Nandi PK, Leclerc E, Nicole J-C, Takahashi M. DNA-induced partial unfolding of prion protein leads to its polymerization to amyloid. J. Mol. Biol. 2002;322:153–161. doi: 10.1016/s0022-2836(02)00750-7. [DOI] [PubMed] [Google Scholar]
- 5.Deleault NR, Lucassen RW, Supattapone S. RNA molecules stimulate prion protein conversion. Nature. 2003;425:717–720. doi: 10.1038/nature01979. [DOI] [PubMed] [Google Scholar]
- 6.Nandi PK, Nicole J-C. Nucleic acid and prion protein interaction produces spherical amyloids which can function in vivo as coats of spongiform encephalopathy agent. J. Mol. Biol. 2004;344:827–837. doi: 10.1016/j.jmb.2004.09.080. [DOI] [PubMed] [Google Scholar]
- 7.Deleault NR, Geoghegan JC, Nishina K, Kascsak R, Williamson BA, Supattapone S. Protease-resistant prion protein amplification reconstituted with partially purified substrates and synthetic polyanions. J. Biol. Chem. 2005;280:26873–26879. doi: 10.1074/jbc.M503973200. [DOI] [PubMed] [Google Scholar]
- 8.Lima LMTR, Cordeiro Y, Tinoco LW, Marques AF, Oliveira CLP, Sampath S, Kodali R, Choi G, Foguel D, Torriani I, et al. Structural insights into the interaction between prion protein and nucleic acid. Biochemistry. 2006;45:9180–9187. doi: 10.1021/bi060532d. [DOI] [PubMed] [Google Scholar]
- 9.King DJ, Safar JG, Legname G, Prusiner SB. Thioaptamer interactions with prion proteins: sequence-specific and non-specific binding sites. J. Mol. Biol. 2007;369:1001–1014. doi: 10.1016/j.jmb.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 10.Deleault NR, Harris BT, Rees JR, Supattapone S. Formation of native prions from minimal components in vitro. Proc. Natl Acad. Sci. USA. 2007;104:9741–9746. doi: 10.1073/pnas.0702662104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geoghegan JC, Valdes PA, Orem NR, Deleault NR, Williamson RA, Harris BT, Supattapone S. Selective incorporation of polyanionic molecules into hamster prions. J. Biol. Chem. 2007;282:36341–36353. doi: 10.1074/jbc.M704447200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nieto C, Giraldo R, Fernández-Tresguerres E, Díaz R. Genetic and functional analysis of the basic replicon of pPS10, a plasmid specific of Pseudomonas isolated from Pseudomonas syringae pv. savastanoi. J. Mol. Biol. 1992;223:415–426. doi: 10.1016/0022-2836(92)90661-3. [DOI] [PubMed] [Google Scholar]
- 13.Giraldo R, Fernández-Tresguerres ME. 20 years of the pPS10 replicon: insights on the molecular mechanism for the activation of DNA replication in iteron-containing bacterial plasmids. Plasmid. 2004;52:69–83. doi: 10.1016/j.plasmid.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 14.Díaz-López T, Lages-Gonzalo M, Serrano-López A, Alfonso C, Rivas G, Díaz-Orejas R, Giraldo R. Structural changes in RepA, a plasmid replication initiator, upon binding to origin DNA. J. Biol. Chem. 2003;278:18606–18616. doi: 10.1074/jbc.M212024200. [DOI] [PubMed] [Google Scholar]
- 15.Díaz-López T, Dávila-Fajardo C, Blaesing F, Lillo MP, Giraldo R. Early events in the binding of the pPS10 replication protein RepA to single iteron and operator DNA sequences. J. Mol. Biol. 2006;364:909–920. doi: 10.1016/j.jmb.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 16.Giraldo R, Andreu JM, Díaz-Orejas R. Protein domains and conformational changes in the activation of RepA, a DNA replication initiator. EMBO J. 1998;17:4511–4526. doi: 10.1093/emboj/17.15.4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giraldo R, Fernández-Tornero C, Evans PR, Díaz-Orejas R, Romero A. A conformational switch between transcriptional repression and replication initiation in the RepA dimerization domain. Nat. Struct. Biol. 2003;10:565–571. doi: 10.1038/nsb937. [DOI] [PubMed] [Google Scholar]
- 18.Giraldo R. Defined DNA sequences promote the assembly of a bacterial protein into distinct amyloid nanostructures. Proc. Natl Acad. Sci. USA. 2007;104:17388–17393. doi: 10.1073/pnas.0702006104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Christensen DD. Alzheimer's disease: progress in the development of anti-amyloid disease-modifying therapies. CNS Spectr. 2007;12:113–123. doi: 10.1017/s1092852900020629. [DOI] [PubMed] [Google Scholar]
- 20.Necula M, Kayed R, Milton S, Glabe CG. Small molecule inhibitors of aggregation indicate that amyloid β oligomerization and fibrillization pathways are independent and distinct. J. Biol. Chem. 2007;282:10311–10324. doi: 10.1074/jbc.M608207200. [DOI] [PubMed] [Google Scholar]
- 21.Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comp. Chem. 1998;19:1639–1662. [Google Scholar]
- 22.Wiseman T, Willston S, Brants JF, Lin LN. Rapid measurements of binding constants and heats of binding using a new titration calorimeter. Anal. Biochem. 1989;179:131–137. doi: 10.1016/0003-2697(89)90213-3. [DOI] [PubMed] [Google Scholar]
- 23.Jelesarov I, Bosshard HR. Isothermal titration calorimetry and differential scanning calorimetry as complementary tools to investigate the energetics of biomolecular recognition. J. Mol. Recogn. 1999;12:3–18. doi: 10.1002/(SICI)1099-1352(199901/02)12:1<3::AID-JMR441>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 24.García de Viedma D, Giraldo R, Ruíz-Echevarría MJ, Lurz R, Díaz-Orejas R. Transcription of repA, the gene of the initiation protein of the Pseudomonas plasmid pPS10, is autorregulated by interactions of the RepA protein at a symmetrical operator. J. Mol. Biol. 1995;247:211–223. doi: 10.1006/jmbi.1994.0134. [DOI] [PubMed] [Google Scholar]
- 25.McGovern SL, Caselli E, Grigorieff N, Shoichet BK. A common mechanism underlying promiscuous inhibitors from virtual and high-throughput screening. J. Med. Chem. 2002;45:1712–1722. doi: 10.1021/jm010533y. [DOI] [PubMed] [Google Scholar]
- 26.Cohen T, Frydman-Marom A, Rechter M, Gazit E. Inhibition of amyloid fibril formation and cytotoxicity by hydroxyindole derivatives. Biochemistry. 2006;45:4727–4735. doi: 10.1021/bi051525c. [DOI] [PubMed] [Google Scholar]
- 27.Nakamura A, Wada C, Miki K. Structural basis for regulation of bifunctional roles in replication initiator protein. Proc. Natl Acad. Sci. USA. 2007;104:18484–18489. doi: 10.1073/pnas.0705623104. [DOI] [PMC free article] [PubMed] [Google Scholar]