Abstract
U1 interference (U1i) is a novel method to block gene expression. U1i requires expression of a 5′-end-mutated U1 snRNA designed to base pair to the 3′-terminal exon of the target gene's pre-mRNA that leads to inhibition of polyadenylation. Here, we show U1i is robust (≥95%) and a 10-nt target length is sufficient for good silencing. Surprisingly, longer U1 snRNAs, which could increase annealing to the target, fail to improve silencing. Extensive mutagenesis of the 10-bp U1 snRNA:target duplex shows that any single mismatch different from GU at positions 3–8, destroys silencing. However, mismatches within the other positions give partial silencing, suggesting that off-target inhibition could occur. The specificity of U1i may be enhanced, however, by the fact that silencing is impaired by RNA secondary structure or by splicing factors binding nearby, the latter mediated by Arginine-Serine (RS) domains. U1i inhibition can be reconstituted in vivo by tethering of RS domains of U1-70K and U2AF65. These results help to: (i) define good target sites for U1i; (ii) identify and understand natural cellular examples of U1i; (iii) clarify the contribution of hydrogen bonding to U1i and to U1 snRNP binding to 5′ splice sites and (iv) understand the mechanism of U1i.
INTRODUCTION
Technologies to silence specific vertebrate genes have rapidly developed for studying the function of individual genes and hold great promise as molecular therapies. The leading technology is RNA interference (RNAi), which can inhibit gene expression 5- to 20-fold. U1i (U1 small nuclear RNA–U1 snRNA- interference) is a relatively new addition to the gene silencing tool kit that gives impressively high levels (up to 1000-fold) of silencing of reporter genes (1–3) and high levels (20-fold) of endogenous genes (2,3). U1i has recently been shown to have efficacy against HIV replication (4) and also works in animal models (Abad,X. and Puri,F., unpublished data) and thus holds great promise for knockdown studies and gene therapy applications. U1i is based on the finding that a U1 snRNP bound to the 3′-end of a pre-mRNA can inhibit pre-mRNA 3′-end processing and therefore the gene's expression. The processing of pre-mRNA to become mature mRNA is an obligatory step in the expression of eukaryotic protein-encoding genes (5). Nearly all metazoan pre-mRNAs undergo 5′ end capping, splicing to remove introns and join exons, and 3′ end processing, where the poly(A) tail is added by a cleavage and polyadenylation reaction that requires a poly(A) signal A(A/U)UAAA and a downstream GU-rich sequence that flanks the poly(A) site. The mature mRNA is then exported to the cytoplasm where it can be translated.
The U1 small nuclear ribonucleoprotein (U1 snRNP) is a constitutive splicing factor and, for certain viral genes, can regulate 3′-end processing. In humans, U1 snRNP is comprised of the 164-nt long U1 snRNA bound by 10 proteins: seven Sm proteins and three U1 snRNP-specific proteins U1A, U1C and U1-70K (Supplementary data Figure S1). U1 snRNP functions in pre-mRNA splicing by choosing the donor exon–intron boundary via a base pairing interaction between nts 2–11 of U1 snRNA and the 5′ splice site sequence (ss) (6). The most definitive experiment to show this interaction involved expression of 5′-end-mutated U1 snRNAs that restored splicing activity to a mutated 5′ss only when nts 2–11 of the U1 snRNA were complementary to the mutated 5′ss (7–11). Of the ∼300 000 human natural 5′ss sequences most have a modest complementarity with U1 snRNA with the mean being 6/10 nt. Thus, many factors have been described that regulate U1 snRNP binding to the 5′ss (12), such as the nuclear cap binding complex (CBC), T-cell intracellular antigen 1 (TIA-1) or SR proteins that bind splicing enhancers or silencers. CBC binds the nuclear cap at the 5′end of the pre-mRNA and increases the binding of U1 snRNP to the first 5′ss (13). TIA-1, a factor linked to translation control binds the RNA downstream of some 5′ss sequences and increases U1 snRNP binding to a weak 5′ss by direct interaction with U1C (14,15). SR proteins bind sequences close to the 5′ss and affect U1 snRNP binding by interaction with U1-70K leading to exon skipping or inclusion (16,17).
Aside from this well-studied splicing function, U1 snRNP can also act as a potent inhibitor of gene expression by inhibiting pre-mRNA 3′ end formation. The expression of late genes of certain papillomaviruses was the first identified natural occurring example of this inhibitory activity of U1 snRNP (2). Inhibition requires U1 snRNP to base pair to a target 5′ss-like sequence located in the 3′ terminal exon of the papillomavirus mRNA. We call such 3′-terminal exon sequences U1-binding sites so as to distinguish them from 5′ss sequences as they are functionally different. The same U1 snRNA nts 2–11 that are used in 5′ss recognition are also used to base pair to the papillomavirus U1-binding site. Subsequent to its discovery in papillomaviruses, a number of studies established key aspects of the inhibitory mechanism. After U1 snRNP binding to the U1-binding site, the U1-70K component of the U1 snRNP directly binds to and inhibits the polyadenylation activity of nuclear poly(A) polymerase, the enzyme that catalyzes addition of the poly(A) tail (18). This was shown both in vitro and in vivo as exogenous U1 snRNAs that lack the U1-70K binding site fail to inhibit polyadenylation (1,4). Thus, inhibited pre-mRNA is cleaved at the 3′-end but it is not polyadenylated. Without a poly(A) tail, the pre-mRNA fails to mature and is rapidly degraded in the nucleus leading to reduced levels of that gene's mRNA. Up to very recent, natural U1-binding sites had only been found in papillomaviruses. However, we recently identified the first example of a mammalian gene having a natural U1-binding site in the 3′ terminal exon (45). Although this site matches the consensus sequence, and so should be constitutively active, its activity is influenced by two types of flanking sequences, one that represses via an RNA secondary structure and the other that stimulates via binding a trans-acting factor. Thus, the inhibitory activity of strong U1-binding sites can be readily regulated and so represent an additional mechanism for the cell to regulate the biosynthesis and activity of mRNA.
This inhibitory activity of U1 snRNP forms the basis of the U1i silencing technology, which is designed to mimic what has been described in papillomavirus. Expression of an exogenous U1 snRNA, whose 5′-end has been modified to base pair with a target mRNA sequence, inhibits target mRNA expression by inhibiting polyadenylation (1,3,19). The specificity of inhibition is underscored by the fact that U1i does not affect expression of certain histone mRNAs that lack a poly(A) tail. The 3′-end processing of most of the replication-dependent histone mRNAs utilize a specialized stem–loop signal to produce a mature mRNA 3′ end (20). The inhibitory mechanism of U1i is specific to the poly(A) signal as inhibition was lost when the target gene's poly(A) signal was replaced by such a histone 3′ end formation signal (3). The inhibitory affect of U1 snRNP on poly(A) polymerase would lead one to expect that an optimal target sequence should be close to the site of poly(A) tail addition. Surprisingly, this is not the case as strong inhibition was seen even when the target site was moved far (up to 1190 nt) upstream of the poly(A) signal. However, inhibition was lost when the U1 target site was placed upstream of an intron (3). This and other reports led to the realization that the U1 target site needs to be in the 3′-terminal exon to inhibit polyadenylation. This makes the U1i system distinct from traditional inhibitory antisense technologies or RNAi where the target site can be in any part of the mRNA and the mechanism involves degradation of mature cytoplasmic mRNA or translation inhibition (21–23).
The rules that govern U1i were first studied in reporter genes where inhibition is conferred by insertion into the terminal exon of a consensus U1 target site that binds endogenous U1 snRNA (1–3). Insertion of a single U1 target site typically gave high inhibitory activities in the range of 15- to 30-fold and insertion of two or more U1 target sites gave synergistic, enhanced inhibitory activity up to 1000-fold. Such high inhibitory levels are unprecedented. The application of U1i to silence endogenous genes requires expression of a 5′-end-mutated U1 snRNA and typically gives 10- to 20-fold inhibitory levels. Expression of multiple 5′-end mutated U1 snRNAs that target different parts of the terminal exon of the same gene also gives synergistic enhanced inhibition. Also, inhibition works in both transiently and stably transfected cells.
In spite of the great potential of U1i, the success of this technique has been hampered by the paucity of information regarding the nature of the target sites and the modifications that can be done to the U1 snRNA that result in good inhibitory molecules. In this work, we study the effect on silencing efficiency of varying the length of the U1 snRNA:target site duplex and the presence of mismatches in the duplex. We also analyze the effect of inhibition when the target site is either close to or partially within structured regions of the mRNA or is near sequences bound by factors involved in constitutive or alternative splicing. Finally, we dissect the partners involved in U1i. Removal of U1-70K protein results in an inactive U1i complex, which can be reactivated by tethering to the complex RS domain of U1-70K or of the splicing factor U2AF65. These results provide a new insight into the mechanism by which U1i works, guide the design of good U1i inhibitory molecules and are important for predicting naturally occurring U1-binding sites and the factors that regulate them. Also, our results may help to understand U1 snRNA binding to the pre-mRNA during splicing as they serve to highlight the importance of hydrogen bonding to allow 5′ss recognition by U1 snRNP.
MATERIALS AND METHODS
Cell lines and transfections
HeLa cells were obtained from the ATCC and cultured in DMEM medium, supplemented with 10% FBS and 1% penicillin–streptomycin at 37°C in a 5% CO2 atmosphere. All cell culture reagents were obtained from Invitrogen. Plasmids were transfected using calcium phosphate as described (24) or Polyfect (Qiagen) as per manufacturer's instructions.
Plasmids
Either pGL2 or pGL3-Promoter plasmids (Promega) were used as a firefly luciferase transfection control. All the Renilla luciferase expressing plasmids used in this work were derived from pRL-SV40 (Promega). The pRL/87wtU1 and pRL/145wtU1 have already been described and were previously called pSV/87WT and pSV/145WT, respectively (3). The Renilla plasmids in Figures 1, 2, 5 and 6 with U1-binding site insertions were made by ligation of double-stranded oligonucleotides (Sigma) into the XbaI–NotI site of pRL-SV40 (Promega). The Renilla plasmids in Figures 3 and 4 with U1-binding site insertions were derived from pL3 (3) that had an Adenovirus L3-derived 3′UTR and poly(A) signal sequences in place of the SV40 3′UTR and poly(A) signal sequences. The Renilla plasmid with two MS2 binding sites in Figure 7D and E are as previously described (46). MS2 plasmids express MS2 or MS2 fused to the RS domain of ASF/SF2 (MS2/ASF/SF2), U2AF65 (MS2/U2AF65), U1-70K (MS2/70K) or the mutated RS domain of ASF/SF2 (MS2/mutASF/SF2) or U2AF65 (MS2/mutU2AF65) and have been described previously (46). A wild-type U1 snRNA (wtU1) expression plasmid was constructed by inserting a 600-bp human U1 snRNA gene, including promoter and termination sequences, into the BamH1 site of pGem3Z+. All the U1 snRNA expression plasmids had a mutation in stem–loop 3 as previously described (1,3) that allowed for measuring expression of exogenous U1 snRNA without detection of endogenous U1 snRNA. The wtU1 plasmid was used to construct plasmids expressing 5′-end modified U1 snRNAs such as mtU1/+0 (Figures 1 and 6), 8bpmtU1 (Figure 6), mtU1/+0/MS2 (Figure 7) or mtU1 with the indicated extensions in the 5′-end of U1 snRNA (Figure 1). To this end, base paired oligonucleotides (Sigma) designed to contain the sequences indicated in Figures 1 and 6, were ligated into the BglII–BclI site of the wtU1 plasmid. To construct mtU1/+0/MS2, we used a wtU1 plasmid in which the stem–loop 1 sequences GGAGATACCATGATCAACGAAGGTGGTTTTCC of human U1 snRNA were replaced by an MS2 binding sequence CCTGATACACCATCAGGGTTCAGG. All the clones have been verified by sequencing in an ABI Prism 310 genetic analyzer (Applied Biosystems). Plasmid DNA was purified with a Maxiprep kit (Qiagen) before transfection.
Figure 1.
Inhibitory activity of U1 snRNAs having extended 5′ends. (A). Exogenous 5′-end mutated U1 snRNA inhibits the expression of the targeted Renilla reporter in a dose-dependent manner. HeLa cells were cotransfected with a Renilla pRL/87mtU1 reporter that contains a mtU1 binding site at position –87 and variable amounts (1, 2 or 3 µg) of mtU1/+0 plasmid that expresses a 5′-end mutated U1 snRNA designed to base pair to the mtU1-binding site as indicated. ‘+0’ serves to indicate a normal length 5′ end as opposed to an extended 5′ end. As a control, a plasmid expressing wtU1 was used in place of the mtU1/+0 plasmid. In all cases, a firefly plasmid was also cotransfected for normalization purposes. The inhibitory activity of the mtU1/+0 plasmid was calculated by dividing the normalized Renilla activity of the transfection with the pRL/87mtU1 + wtU1 plasmids by the normalized Renilla activity of the transfection with the pRL/87mtU1 + mtU1/+0 plasmids. The wtU1 plasmid did not affect expression of pRL/87mtU1 and therefore had an inhibitory value set to 1.0. The standard deviations were calculated from three independent experiments. (B) 5′ end extended U1 snRNAs do not increase inhibitory activity. The plasmid that expresses mtU1/+0 was used as a parental vector to construct plasmids that express U1 snRNA extended at the 5′-end +1, +2, +3, +4, +5, +6 and +15 nt (mutU1/+1, mutU1/+2, etc.). These extensions increase the length of the duplex formed between pRL/87mtU1 pre-mRNA and the mtU1-derived snRNA. The sequence of the 5′ end of the U1 snRNA and the duplex length is indicated for each case. As described in (A), all plasmids were transfected along with pRL/87mtU1 and firefly luciferase and their relative inhibitory activities were calculated. The inhibitory activities are derived from five different experiments. Standard deviations are not shown but were <25% in each case. (C) 5′-end extended snRNAs are stably expressed. A primer extension reaction was performed with no RNA (lane 1) or with total RNA isolated from cells transfected with the wtU1 expressing plasmid (lane 2) or the mtU1-derived plasmids from (B) as indicated (lane 3–10). The 32P-end-labeled oligo used recognizes both endogenous wtU1 and all mtU1-derived snRNAs, as indicated to the right of the figure. The samples were separated by denaturing PAGE and visualized by autoradiography. The product obtained with mtU1/+0 snRNA in lane 3 comigrates with the product of endogenous U1 snRNA. The product of mtU1/+1 snRNA in lane 4 is difficult to visualize because it would be only 1-nt longer than the product of endogenous U1 snRNA.
Figure 2.
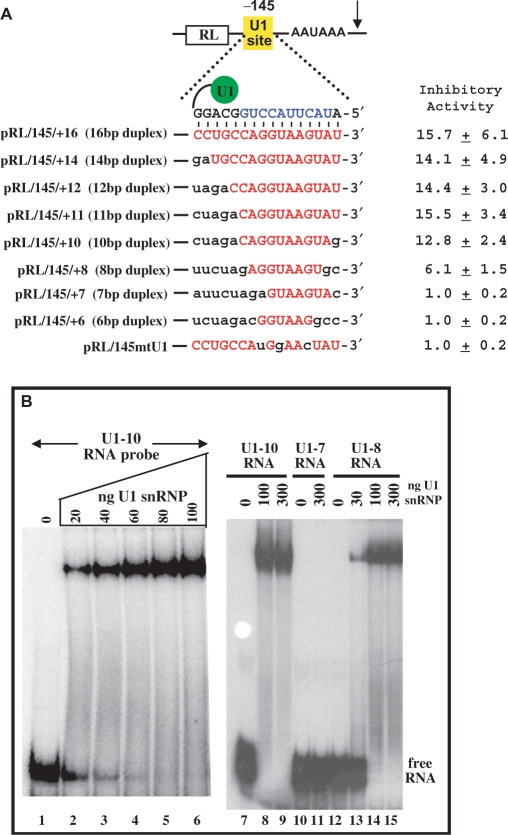
Effect of U1-binding site:U1 snRNA duplex length on inhibition. (A) Shown is a series of Renilla luciferase plasmids with different U1-binding sites cloned at 145 nt from the poly(A) signal (pRL/145/x). Shown in red are nucleotides from the U1-binding site able to bind endogenous U1 snRNA. The U1-binding site:U1 snRNA duplex length is indicated for each case. As was done in the Supplementary Data Figure S1, HeLa cells were transfected with these plasmids along with a firefly luciferase plasmid for normalization purposes. Inhibitory activities were calculated as in the Supplementary Data Figure S1 and the bar graph summarizes five independent experiments. The pRL/145mtU1 is the reference control plasmid that matches pRL/145/+16 except for three point mutations in the U1-binding site. (B) An electrophoretic mobility shift assay (EMSA) was used to detect binding of purified U1 snRNP to 0.03 pmol 32P-radiolabeled RNAs with various types of U1-binding sites. EMSA conditions were as previously described (18). The RNAs are all matching except for differences in the U1-binding site sequence. Lanes 1–9 contain 32P labeled U1-10 RNA that has a 10-nt wt U1-binding site. Lanes 10 and 11 contain U1-7 RNA that matches U1-10 RNA except the U1-binding site is 7-nt long. Lanes 12–15 contain U1-8 RNA that matches U1-10 RNA except the U1-binding site is 8-nt long. The amounts of purified U1 snRNP added are indicated (note 100 ng U1 snRNP = 0.3 pmol). The purification of U1 snRNP is described in the Supplementary Data Figure S2. The experiment was repeated ×5, quantitated by phosphorimagery and a kDa of 4 +/− 1.5 nM was calculated for the U1 snRNP:U1-10 RNA complex. The U1-8 RNA bound about 3× weaker than the U1-10 RNA and no detectable binding to the U1-7 RNA was observed under these conditions.
Figure 5.
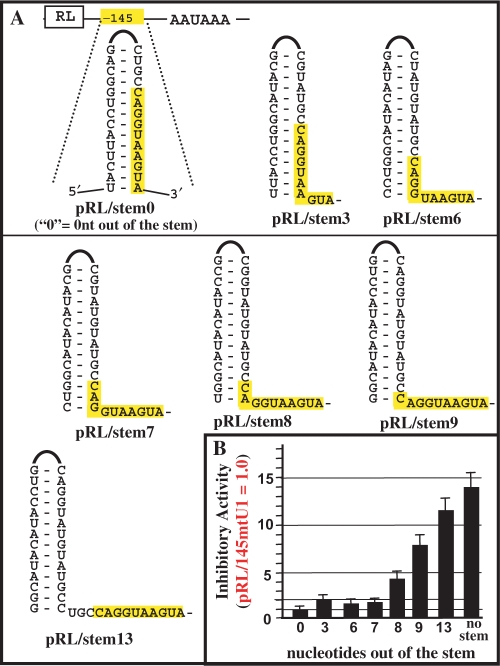
Effect of secondary structure on U1-binding site inhibition. (A) The pRL/145/stem0 plasmid has a 13-nt wtU1-binding site (the canonical 10 nt are highlighted in yellow) completely occluded in a stem–loop sequence. The ‘0’ indicates 0 nt of the U1-binding site should be found outside of the stem. As diagrammed, a collection of plasmids were made and tested that match pRL/145/stem0 except the U1-binding site increasingly moves out of the stem. The number below each stem–loop structure indicates the number of U1-binding site nucleotides that should be found outside of the stem (3, 6, 7, 8, 9 or 13). (B) The plasmids were analyzed and graphed as indicated in Figure 3. Error bars indicate standard deviations of four different experiments.
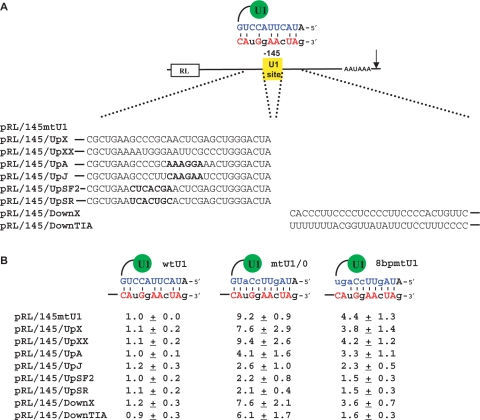
Figure 6.
Splicing regulatory sequences interfere with U1 snRNP's poly(A) site inhibitory activity. (A) Plasmid pRL/145mtU1, whose binding to endogenous U1 snRNP is schematized at the top of the figure, served as a parental plasmid to insert specific sequences upstream or downstream of the U1 target site, as indicated. Sequences chosen are control sequences (UpX, UpXX or Down X), or sequences A, J, SF2, SR and TIA-1, which bind unknown factors (A and J), SF2/ASF, SRp40 and TIA-1, respectively, in the nucleotides shown in bold. The rest of the sequence has been included to keep the context that is known to affect splicing activity. (B) Each Renilla plasmid was transfected into HeLa cells along with a firefly luciferase expressing plasmid as a transfection control and a U1 snRNA expression plasmid: either wtU1, or mtU1/0 (same as in Figure 1) that should form a 10-bp duplex or 8bpmtU1 that should form an 8-bp duplex. Indicated at the top of the panel is the predicted duplex formed between the exogenous U1 snRNA and the U1 target site sequence. Luciferase activity was measured and normalized to calculate the inhibitory activity in each case. The results show the average of three independent experiments.
Figure 3.
Analysis of single-point mutations of the U1-binding site. A saturation mutagenesis analysis was performed where all 30 possible single-point mutations were introduced in the 10-nt wtU1-binding site of the pRL/180wtU1 plasmid shown in the Supplementary Data Figure S1. Each plasmid was cotransfected with the firefly luciferase control into HeLa cells and inhibitory activities were calculated as described in the Supplementary Data Figure. S1. The results are plotted in a bars graph where error bars indicate standard deviations of five independent experiments. The sequence of the wtU1-binding site and its base pairing with the 11 nt of the 5′-end of U1 snRNA (blue font) is schematized above the graph. Each dotted line indicates the mutation (in red font) which is positioned above its corresponding bar graph representing the inhibitory activity of that mutation. The inhibitory activity of the reporter with a wtU1-binding site is shown in green to the left of the graph. The 6G and 7G single mutants were combined to give the 6G/7G double mutant whose activity is shown on the far right.
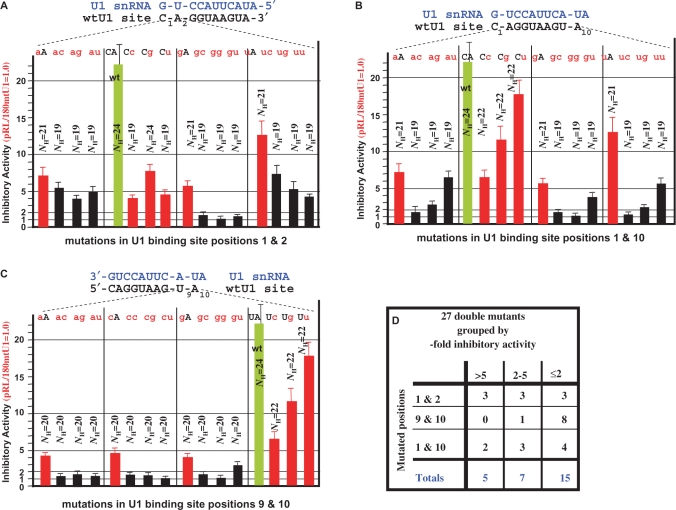
Figure 4.
Analysis of U1-binding sites with double-point mutations at positions 1 and 2, 1 and 10 or 9 and 10. A saturation mutagenesis analysis was performed where all possible double mutations were introduced at positions 1 and 2 (A) 1 and 10 (B) and 9 and 10 (C) of the 10-nt wtU1-binding site of the pRL/180wtU1 plasmid shown in the Supplementary Data Figure S1. The mutations were analyzed and graphed as in Figure 3. The sequence of the wtU1-binding site and its base pairing with the 11 nt of the 5′-end of U1 snRNA (blue font) is schematized above the graph. Each pair of letters indicates a particular mutant where the lowercase red letters correspond to a mutation, while the uppercase black letters match the wtU1-binding site. Each pair of letters is positioned above its corresponding bar graph representing the inhibitory activity of that mutated U1 site. Also indicated are the NH values that are defined as the number of continuous hydrogen bonds (see Discussion section). To facilitate comparison we included the single-point mutations from Figure 2 as red histograms. All double mutants (totals) or double mutants from (A) (1 and 2), (B) (1 and 10) and (C) (1 and 10) were classified according to their good, low or no inhibitory activity (>5-, 2–5- and ≤2-fold inhibition, respectively). Shown is the number of double mutants in each group (D).
Figure 7.
Analysis of the role of RS domains in U1i. (A) Schematic of mutU1/+0/MS2, the modified U1 snRNA used to test the inhibitory activity of various MS2 fusion proteins. mutU1/+0/MS2 is identical to U1 snRNP shown in the Supplementary Data Figure S1A but loop 1 has been replaced by an MS2-binding sequence and the 5′-end binds a mutated sequence in the 3′ terminal exon of the Renilla reporter plasmid as shown. (B) Analysis of the expression of MS2 fusion proteins. HeLa cells were transfected with the plasmids that express a control protein, MS2 alone or MS2 fused to the RS region of U1-70K (MS2/70K) or the RS region of U2AF65 (MS2/U2AF65) and extracts were collected at 48 h posttransfection. MS2 expression was evaluated in the extracts by western blot analysis. (C) Reconstitution of functional U1i complexes by tethering RS domains to the loop 1 of U1 snRNA. HeLa cells were cotransfected with three plasmids: (i) a Renilla construct that binds mutU1/+0/MS2, (ii) a plasmid that expresses either a control protein, MS2 alone or MS2 fused to the RS region of U1-70K (MS2/70K) or the RS region of U2AF65 (MS2/U2AF65) and (iii) a plasmid that expresses either mutU1/+0/MS2 or a control U1 snRNA. In all cases, a plasmid expressing firefly luciferase was also cotransfected as a control. Extracts were collected at 48 h posttransfection and luciferase activity was evaluated. All data were normalized to firefly luciferase expression. Renilla expression in the presence of a control U1 snRNA was similar in all cases and was used to calculate the fold inhibition. (D) Design of a Renilla reporter that tethers RS domains upstream of an active wtU1-binding site. The two MS2 stem–loops are 42-nt apart and collectively are 17-nt upstream of the wtU1-binding site. (E) Disruption of the inhibitory activity of the wtU1-binding site by a cotransfected MS2 fusion protein. Shown are the results where the reporter in (D) is cotransfected with either an empty vector ‘no MS2’ or an MS2 fusion expression plasmid that expresses MS2 protein or MS2 fused to RS domains from various SR proteins as indicated. The MS2/mtU2AF65 and MS2/mtASF/SF2 are controls that express a mutated RS domain from U2AF65 or ASF/SF2, respectively. The mutations and the RS domain are as previously described (46). Western blotting was used to confirm that the MS2 fusion proteins were expressed to a similar level (data not shown).
Luciferase activity measurements
Renilla and firefly luciferase activity was quantified using the Dual Luciferase System (Promega) as previously described (24) in a Berthold Luminometer (Lumat LB 9507).
Western blot
The MS2 domain was detected by western blot as previously described (47) using a 1:2500 dilution of anti-MS2 antibody kindly provided by Dr Peter G. Stockley and Gabriella Basnak.
Preparation and analysis of RNA
Total RNA was purified using guanidium thiocyanate as described (25). Primer extensions were done with 6 pmol of ATP γ-labeled oligo 5′-GCCCTGGGAAAACCACCTTCG-3′ incubated with 0.75 μg of total RNA, as described. These conditions were set up to work in a linear range. Samples were loaded onto a 14% polyacrylamide gel and separated by electrophoresis. Gels were dried and exposed to a screen that was developed in a Cyclone phosphorimager (Perkin Elmer).
In vitro binding assay
Electrophoretic mobility shift assay was done as previously described (18) and contained 32P-labeled RNA, 0.7 mM MnCl2, 50 mM KCl, 0.1% Triton X-100, 1 µg total yeast tRNA, 20 mM Tris–HCl (pH 7.5), 3 U RNasin (Promega), 0.1 mM EDTA and 3 µg bovine serum albumin in 15 µl. U1 snRNP was added last and the reaction incubated 5 min at room temperature prior to loading a 6% (60:1) polyacrylamide gel run in Tris–borate–EDTA buffer. Electrophoresis was for 3 h at 20 V/cm. The purification of U1 snRNP from HeLa cells is described in the Supplementary Data Figure S2.
RESULTS
U1i is a robust inhibitory mechanism
To systematically analyze parameters that effect U1i we determined whether inhibition levels would be influenced by the amount of transfected plasmid or time of transfection. To this end, we utilized the dual luciferase reporter system (Promega) where the pRL/180wtU1 Renilla luciferase reporter plasmid was targeted for inhibition by insertion of a 10-nt wild-type U1 snRNA (wtU1) binding site that can base pair endogenous U1 snRNA. A control plasmid (pRL/180mtU1) was made by insertion of a mutant U1 snRNA (mtU1) binding site which cannot bind endogenous U1 snRNA. All Renilla values used in this work were normalized to a cotransfected firefly luciferase plasmid. The fold inhibition by U1i is simply the ratio of the normalized Renilla expression of the mutant plasmid (pRL/180mtU1) to that of the wild-type plasmid (pRL/180wtU1) 9. We varied the amount of transfected plasmid over a 1000-fold range and found that fold inhibition by U1i remained consistently high (Supplementary Data Figure S1). Similarly, a time-course experiment showed that that U1i inhibition was consistently high from 12 to 72 h after transfection (Supplementary Data Figure S1). We also found U1i to give good inhibition when these constructs were transfected in a variety of other human cell lines (for example, K562, DU145, HEK, HEK293, A549, PC3, LnCaP, SK-N-MC, 293F, Jurkat, BHK to list a few), cell lines from other vertebrates (monkey, mouse, rat and chicken), as well as from primary vertebrate cells (data not shown). These results show that the molecular mechanism that allows inhibition is robust over a wide range of expression levels of mRNA, time points and cell types.
5′-end extended U1 snRNAs do not give enhanced inhibition
Next, we wanted to determine whether lengthening the 5′-end of U1 snRNA would improve inhibition. Endogenous U1 snRNA is transcribed in the nucleus, and then transported to the cytoplasm where Sm proteins bind, the 3′ end is processed and the 5′-end is modified with a trimethyl-guanosine (TMG) cap (6). Then, U1 snRNA is re-imported into the nucleus and binds the U1 snRNP-specific proteins to form a mature U1 snRNP. Others and we have previously shown that U1 snRNAs with 5′-end mutations have a similar maturation and are capable of gene expression inhibition. This is achieved by transfection of a wtU1 plasmid that contains the modified U1 snRNA sequence under a U1 snRNA promoter and termination signal. Thus, we expressed a mutant U1 snRNA (called mtU1/+0 snRNA) designed to base pair a mtU1-binding site inserted 87-nt upstream of the poly(A) signal in the pRL/87mtU1 plasmid (Figure 1A). Co-expression of the mtU1/+0 snRNA and the pRL/87mtU1 Renilla reporter resulted in an inhibition of the expression of the reporter of up to 20-fold (Figure 1A). As expected, this inhibition decreased when the amount of the mtU1/+0 snRNA was decreased (Figure 1A). The 20-fold level of inhibition is quite respectable as the mtU1/+0 is expressed at a lower level than endogenous U1 snRNA in transfected cells, which inhibits at a 30-fold level the pRL/87wtU1 plasmid that has a wtU1-binding site.
Having established the level of inhibition by mtU1/+0 snRNA, we were now able to determine whether U1 snRNA 5′ end extensions would increase inhibition. We rationalized that the inhibition obtained from these snRNAs would increase as the base pairing potential to the target sequence increased. Thus, we constructed a series of mtU1/+0 plasmids that express mutant U1 snRNAs with the 5′ end extended in 1-nt increments (from 1 to 6) or 15 nt, which would increase the duplex formed by the 5′-end of U1 snRNA and the target sequence from 15 to 31 nt (Figure 1B). Unexpectedly, most of the 5′ end extended U1 snRNAs were poor inhibitors and some of them had lost inhibitory activity. The same low activity was obtained with other 5′-end extended U1 snRNAs, even when the length of its base pairing to the target mRNA was kept constant or when the U1 snRNA extensions ended in a stem structure to help in stability (data not shown). The only exception to this pattern was the mtU1/+6 snRNA that gave an inhibition level similar to the nonextended mtU1/0 snRNA. Although the basis for the aberrant activity of mtU1/+6 is not clear, we speculate it may be due to antisense affects of the extended U1 snRNA rather than polyadenylation inhibition, as a similar mtU1/+6 construct that only binds 15 nt of the target has low inhibitory activity (data not shown). As shown in Figure 1C, we confirmed that all the 5′ end extended U1 snRNAs were expressed in the cell by primer extension. Northern blotting and an additional primer extension assay using an oligo specific to a sequence tag in stem–loop 3 also demonstrated these U1 snRNAs were expressed at the same level (data not shown). We also confirmed that these U1 snRNAs contained the TMG cap by performing anti-TMG antibody immunoprecipitations and that the mature forms were localized to the nuclear fraction after cell fractionation (data not shown). Thus, we conclude the 5′ end extended U1 snRNAs are expressed, but in general, are poor inhibitors as compared to a normal length U1 snRNA.
A 10-bp duplex between U1 snRNA and its binding site gives near maximal inhibition
The previous experiment led us to think that duplex length may not be an important limiting factor for good inhibition. To analyze this further, we tested a series of reporter constructs where the length of the U1-binding site was changed so as to vary its base pairing potential to endogenous U1 snRNA. This had the advantage that all the data are directly comparable as the inhibitor (endogenous U1 snRNA) is identical for all of the reporter plasmids. In contrast, expression of a 5′-end mutated U1 snRNA has the caveat that some mutations may subtly affect the inhibitory function of U1 snRNA by effecting either U1 snRNP maturation or conformation.
As shown in Figure 2, 6 and 7-nt long U1-binding sites gave no inhibitory activity, whereas an 8-nt long U1-binding site that could base pair to positions 3 to 10 of U1 snRNA gave a 6.1-fold inhibition. Later we show that certain 8-nt long U1-binding sites that base pair to positions 4–11 or 2–9 of U1 snRNA have no or very low (≤2-fold) inhibitory activity. Thus, not all 8-nt U1-binding sites are active. Lengthening the U1-binding site from 8 to 10 nt to make a 10-bp duplex further increased inhibition to 12.8-fold. Lengthening to 11, 12, 14 and 16 nt did not give a significant increase in inhibition over that seen with a 10-nt site. Thus, inhibition is observed with target sites of ≥8 nt and inhibition is near maximal with a 10-nt site. However, one caveat is the 12, 14 and 16-nt long U1 sites are designed to base pair to nts 12–15 of endogenous U1 snRNA that are already base paired within stem 1A as part of an intramolecular base pairing interaction (Supplementary Data Figure S1, panel A). Thus, modifications of nts 12–16 of U1 snRNA could affect stem 1A formation and therefore the activity of U1 snRNP. Therefore, we recommend target site lengths of 10–11 nt.
To determine whether the difference in activities between the 7-, 8- and 10-nt long U1-binding sites reflected different binding affinities to U1 snRNP, we performed in vitro binding assays by incubating HeLa cell-purified U1 snRNP (Supplementary Data Figure S2 for purification of U1 snRNP) with RNA templates containing 7-, 8- and 10-nt long U1-binding sites. As shown in Figure 2B, U1 snRNP is capable of stably binding 8 and 10-nt U1-binding sites but not a 7-nt site. Thus, the differences in inhibitory activity in vivo directly correlate with different affinities to U1 snRNP in vitro.
Certain point mutations in the U1-binding site cause loss of inhibition
Previously, we and others showed that a mutated U1-binding site giving several internal mismatches (1,3) or even a single central-mismatch (26) in the U1-binding site:U1 snRNA duplex caused complete loss of inhibitory activity. However, those studies were limited to just three mutants. To determine how each target position contributes to inhibition, a saturation point mutagenesis was performed on the 10-nt U1-binding site. This would also test whether UU and GU base pairs would be tolerated as they are known to contribute to RNA:RNA helix stability in other contexts (27,28). Figure 3 summarizes the analysis of all 30 possible single-point mutations in the canonical 10-nt wtU1-binding site which were made in an isogenic plasmid background (pRL/180wtU1, see Supplementary Data Figure S1) and analyzed by transient transfection. Inhibitory activities range from 22.3-fold for the wtU1-binding site to 1.0-fold (no activity) for a U1-binding site with five point mutations (Supplementary Data Figure S1). As an arbitrary cut-off point, we chose that any mutant with an inhibitory activity ≤2-fold would be considered to have no activity.
The result shows that U1-binding site positions 3–8 are critical to activity as nearly all point mutations abrogated inhibitory activity. The only two exceptions are mutant 6G (6-fold inhibition) and mutant 7G (11.5-fold inhibition) and notably, both would form GU base pairs (GU pairs) with endogenous U1 snRNA. A double mutant combining 6G with 7G gave no inhibitory activity, indicating there is no tolerance for two tandem internal GU pairs at this position (also shown in Figure 3). Interestingly, a UU pair at either position gave no inhibitory activity underscoring the sensitivity of the duplex to small changes in base pairing potential. Before proceeding, we point out that these Us in U1 snRNA are in fact pseudouridines (ψ). Although it is more accurate to refer to these as G-ψ and U-ψ pairs, prior studies have shown however that a base pair containing a pseudouridine has nearly the same thermal stability and hydrogen-bonding potential as a base pair with uridine (29,30). Thus, we will refer to uridines, rather than pseudouridines at these two positions, a convention also employed by nearly all U1 snRNA-related publications (31,32).
Given that an 8-nt U1 site retains about 40% activity of the 10-nt wtU1 site (Figure 2) we were not surprised that all the single-point mutations in positions 1, 2, 9 and 10 were active (Figure 3). The data follow an interesting pattern that positions 1, 2, 9 and 10 can be placed in a hierarchy of 10 > 1 > 2 > 9 where mutating position 10 is most tolerated (most active) and mutating position 9 is least tolerated (least active). Furthermore, all possible uninterrupted duplexes of 9 bp result in inhibitions ≥5-fold (Figure 3). Thus a continuous uninterrupted helix of 9 bp, as opposed to an interrupted 10-bp helix, is more favorable for inhibition. Similarly to what has been described earlier, mutations C1U, A2G and A10G, which would lead to GU pairings, were best tolerated compared to mutations that disrupt standard Watson–Crick pairings. Also, mutation A2U, which could lead to a UU pairing, was not tolerated. However, another UU pairing could be formed in mutation A10U, which resulted in inhibition similar to the one obtained for the wt U1-binding site. We cannot exclude that this UU pairing is not formed and the first A of U1 snRNA or another part of the U1 snRNP complex is interacting with the 10U mutant. More analysis of the point mutants will be given in the ‘Discussion’ section. As in Figure 2B, EMSAs were done on a subset of the single-point mutants to determine whether their differences in inhibitory activities reflect different binding affinities to purified U1 snRNP. As shown in the Supplementary Data Figure S3, the differences in inhibitory activity in vivo closely correlate with different affinities to U1 snRNP in vitro, indicating that binding affinity plays a key role in the inhibitory activity. To determine whether extended base pairing would increase activity of the partially active mutants we changed position 11 from G to U so that it could base pair to the first A of U1 snRNA. As shown in the Supplementary Data Figure S4, we found the G to U change did not stimulate activity of these partially active mutants suggesting base pairing to the first A of U1 snRNA does not contribute to inhibitory activity. Finally, before proceeding, we point out it was possible the mutagenic analysis would inadvertently produce binding sites either for miRNAs or for proteins that affect mRNA stability or translation. To rigorously rule this out, we would have to analyze each point mutation in detail, a formidable task given the number of mutations. We feel the likelihood of this happening is low as all constructs had the same 3′UTR. Furthermore, we performed a bioinformatic analysis that indicated these point mutations do not inadvertently produce new miRNA-binding sites or binding sites for proteins known to regulate mRNA (data not shown).
Analysis of double-point mutations
Given that single-point mutations to positions 1, 2, 9 and 10 retained significant activity, we proceeded to construct and analyze a series of double mutations involving these positions. Twenty-seven double mutants were analyzed, representing all possible mutations of positions 1 and 2 (Figure 4A), positions 1 and 10 (Figure 4B) and positions 9 and 10 (Figure 4C). As was done in Figure 3, any mutant with an inhibitory activity ≤2-fold was considered inactive. In Figure 4D, we group all 27 double mutants according to their inhibitory activity, namely good inhibition (>5), low inhibition (2–5) or no inhibition (≤2). Given the Figure 2 results that an uninterrupted 8-bp duplex had activity, we were surprised that ∼56% (15 of 27) of the double mutants lacked inhibitory activity (<2-fold) (Figure 4D) and this depended on the composition of the unpaired nucleotides flanking the duplex. For example, all the C1G/A2n double mutants, where ‘n’ means the second position can be C, G or U, gave no inhibitory activity (<2-fold) (Figure 4A). However, all the corresponding C1A/A2n double mutants had inhibitory activity >4-fold (Figure 4A). We do not know the reason why a C1G mutation is more destabilizing than a C1A mutation when located before a mismatch and an 8-nt long target site. Similarly, all the C1n/A10C or C1n/A10G double mutants, where ‘n’ is A, G or U, gave very little inhibitory activity, compared to the C1n/A10U double mutants (Figure 4B). This correlates with the strong inhibition observed for the A10U single mutant (Figure 3) that could reflect a noncanonical binding between the U at position 10 and the U1 snRNP. We also speculate that the results obtained with position 10 may reflect an enhanced accessibility of this terminus of the duplex as compared to the other terminus (position 1) that is contiguous with the rest of the U1 snRNA. Finally, an 8-nt long target site followed by any double mutant at positions 9 and 10 gave no inhibition (Figure 4C). This intolerance for mutation of position 9 is also seen with the single mutant hierarchy 10>1>2>9, where position 9 mutants are the least active (Figure 3). Inhibitory activities that could reflect GU base pairings were less apparent in the analysis of the double-point mutations compared to single-point mutations. The reason may be that GU pairings are less stable when close to a mismatch or do not help to stabilize an 8-bp long duplex (Figure 4C).
The U1-binding site loses inhibitory activity when partially masked by a stem
The previous results showing inhibition with certain 8-nt long target sites as well as targets that would produce a GU pair, led us to think that U1i could be more nonspecific than previously thought. However, the fact that many 5′-end modified U1 snRNAs are not cytotoxic even when stably expressed in cells supports the notion that U1i is specific (1,3,4). Thus, we considered the possibility that the natural environment of target sequences could affect U1i specificity. In fact, it has been previously shown that RNA secondary structure inhibits the splicing activity of a 5′ss by occlusion of U1 snRNP base pairing (10,33). Likewise, we previously showed a fully active U1-binding site would completely lose activity when placed within the stem of a 13-bp stem–loop (3). Importantly, such stem–loop structures had no effect on reporter plasmid expression in the absence of a U1-binding site. To further analyze this U1-binding site occlusion, we systematically and incrementally ‘walked’ a U1-binding site out of a stem structure in order to determine when its activity would be restored (Figure 5). The U1-binding site remained essentially inactive (<2-fold inhibition) when 0, 3, 6 and 7 nt were out of the stem, indicating U1 snRNP is not very effective at disrupting stem structures (Figure 5). Inhibitory activity was observed when eight bases of the U1 site were out of the stem and activity further increased when 9 nt and 13 nt were brought out of the stem. The abrupt gain in activity at eight bases is consistent with the data in Figures 2–4.
Placement of splicing regulatory sequences near a U1-binding site decreases inhibitory activity
There are many examples of splicing enhancer and silencer sequences binding regulatory proteins that control splicing by altering 5′ss recognition by U1 snRNP (12). To determine whether such sequences affect poly(A) site inhibition, we inserted five different regulatory sequences into a reporter containing a target site for an exogenous mtU1 (pRL/145mtU1). Sequences A and J have been described to induce an efficient exon inclusion with several minigenes tested (17) (Figure 6A). Sequences that bind SR proteins SF2/ASF or SRp40 were also chosen that induce efficient exon inclusion or exon skipping, respectively, when located at a specific distance upstream of the U1-binding site (17). Finally, we tested a U-rich sequence taken from a Fas intron that activates TIA-1-dependent recognition of the upstream 5′ss (14). The sequences were cloned upstream (A, J, SF2, SRp40) or downstream (TIA-1) of the mtU1 target site and the spacing and sequence context of these motifs matched the configuration that is known to affect splicing activity. Control sequences of the same length were also inserted upstream or downstream the target site. All plasmids were cotransfected with one of the following U1 snRNA plasmids that express (i) wtU1 snRNA, which should not form a stable duplex with the target site, or (ii) mtU1/+0 snRNA (the same as in Figure 1A) that can form a 10-bp duplex with the target or (iii) 8bpmtU1 snRNA that can form 8 bp with the target site (Figure 6B). A firefly luciferase expressing plasmid was also transfected to normalize luciferase activity. The results indicate that the presence of regulatory sequences has no effect on Renilla luciferase expression per se (Figure 6B, left column). However, when the mtU1/+0 snRNA is expressed, all the upstream regulatory sequences decrease inhibition mediated by this snRNA (Figure 6B, middle column). This result was expected with the sequences bound by SRp40 but not for sequences A, J or SF2, which promote exon inclusion and so would be expected to help recognition of the target site. The downstream regulatory sequence did not significantly affect inhibitory activity of the U1 snRNA. The effects were specific as insertion of control sequences had no effect. In agreement with what has been shown in Figure 4, expression of 8bpmtU1, which forms a shorter duplex with the U1 target site, shows a decreased inhibition compared to the mtU1/+0 snRNA that can form a 10-bp duplex (Figure 6B). To determine whether any of the sequences could help to increase the inhibitory effect observed with a suboptimal duplex we tested the 8bpmtU1 snRNA that only forms an 8-bp duplex. Given that TIA-1 has been described to help U1 snRNP binding to weak 5′ss we expected it could help stabilize the 8bpmtU1 snRNA. Surprisingly, this was not the case as TIA-1 sequences and all the other regulatory sequences tested decrease the inhibition observed with 8bpmtU1.
Analysis of the effect of RS domains on the molecular mechanism of U1i
The experiments shown in Figure 6 indicate that splicing factors such as SR proteins decrease silencing efficiency even if they have been described to increase U1 snRNP binding to a 5′ss target sequence. A possible explanation of this apparent contradiction would be that SR proteins help U1 snRNP binding by interaction with the RS domain of U1-70K and, therefore, impede interaction of U1-70K with other factors essential for U1i mediated inhibition of polyadenylation. In fact, U1 snRNP inhibition of papillomavirus late gene expression requires that the RS domain of U1-70K bind the carboxy-terminus of poly(A) polymerase (18). A similar molecular mechanism could be responsible for U1i-mediated inhibition. It has been previously shown that loop 1 of U1 snRNA, where U1-70K binds, is required for inhibition (1,4). To test if the RS domain of U1-70K mediates U1i, we assayed the inhibition obtained with a modified U1 snRNP in which loop 1 has been replaced by an MS2-binding loop (mutU1/+0/MS2) (Figure 7A). As expected, mutU1/+0/MS2 by itself does not support inhibition (Figure 7C). To reconstitute a functional mutU1/+0/MS2, we co-expressed MS2 alone or fused to the RS domain of U1-70K (MS2/70K) or the RS domain of U2AF65 (MS2/U2AF65). Western blotting confirmed expression of the MS2-derived proteins (Figure 7B). Expression of MS2 fused to the RS domains of U1-70K or U2AF65, but not MS2 alone, recovered inhibition with mutU1/+0/MS2 (Figure 7C). This demonstrates that not only the RS domain of U1-70K, but also other RS domains are involved in U1i. This result agrees with the possibility that the molecular mechanism that allows U1i would also require that the RS domain of 70K interacts and inhibits poly(A) polymerase.
Given that SR proteins help U1 snRNP to bind to 5′ splice sites by interaction with the RS domain of U1-70K, they could impede interaction of the RS domain of U1-70K with poly(A) polymerase. Support for such a model comes from the fact that U1-70K's RS-interacting domain and its poly(A) polymerase interacting and inhibitory domain overlap (18,36,46). If the model is true, then we would expect the RS domain of SR proteins to interfere with silencing. To test this, we tethered specific RS domains to the same positions upstream of an active wt U1-binding site by using an MS2 tethering assay. In the absence of MS2 protein expression (the ‘no MS2’ bar graph), the MS2-binding site loops did not effect the activity of the U1-binding site. Expression of MS2 protein with no RS domains also had no effect. In contrast, expression of an MS2 fused with the RS domains of either U2AF65 of ASF/SF2 disrupted the activity of the U1-binding site. This disruption is specific as expression of a mutated RS domain of U2AF65 and of ASF/SF2 had no effect. These results indicate that the RS domain of SR proteins alone is responsible for blocking U1i. To confirm that these MS2-derived proteins are specifically binding to the pre-mRNA substrates in vivo, we made nuclear extract from UV-crosslinked transfected cells, and did immunoprecipitation assays (IP)s with an anti-MS2 antibody as previously described (46). We found the MS2-derived proteins specifically associated with pre-mRNAs containing the wt MS2-binding site as compared to those with a mutated MS2-binding site (data not shown). Thus, we conclude tethering of the SR domains of U2AF65 and ASF/SF2 near to a U1-binding site can disrupt U1i.
DISCUSSION
In this work, we analyze several parameters that affect U1 snRNA-mediated inhibition of gene expression that forms the basis of U1i. Inhibition is robust as expression of both abundant and rare mRNAs in different cell lines are strongly repressed (Supplementary Data Figure S1, and data not shown). In the design of U1i experiments, we recommend modifying the first nucleotides of U1 snRNA (positions 1–11) and avoiding 5′ end extensions of this sequence (Figure 1 and data not shown). It is unclear to us why U1 snRNAs with 5′-extensions are not capable of efficient silencing as they contain the expected TMG cap structure, localize to the nucleus, are bound by Sm proteins and accumulate in the cell to similar levels as U1 snRNAs with a regular length 5′ end. Interestingly, a recent report described natural U1-like snRNAs with longer 5′-ends that are expressed ubiquitously and whose function has not been described yet (34).
Lessons for naturally occurring U1-binding sites
Base pairing of U1 snRNA to the target site is essential for inhibition of the target mRNA. We have studied carefully the influence of the length and the effect of mismatches in the duplex formed by U1 snRNA and the target site. The results are not only interesting for U1i but are also important in predicting and understanding U1-binding sites that naturally occur in the 3′ terminal exon of vertebrate genes. Recently, we used a bioinformatic analysis followed by experimental verification to identify examples of human genes with U1-binding sites in the 3′ terminal exon. A subset of these had the U1-binding site conserved in the 3′ terminal exon of the vertebrate homolog. We then examined in experimental detail a few of these human genes with conserved terminal-exon U1-binding sites and found the U1-binding sites are indeed active in repressing expression (ref. 45 and Goraczniak,R. and Gunderson,S.I., unpublished data). The bioinformatic analysis was based on identifying matches to near-consensus U1-binding sites, namely MAGGUAAGU (M = A or C) and NAGGUAAGUA. Thus we excluded any hits that had an internal mismatch. However, given that positions 6 and 7 can be Us and still retain activity the bioinformatic analysis will have to be expanded to include 6U and 7U which will undoubtedly turn up additional genes with conserved U1-binding sites. Furthermore, such future bioinformatic analysis will also scan for potential secondary structure that is sufficient to strongly impair activity (Figure 5). Indeed, one of the genes we studied in experimental detail (45) has its U1-binding site trapped within a phylogenetically conserved base pairing interaction that base pairs to 5 of 10 nt of the U1-binding site, just sufficient, as per Figure 5, to inhibit its activity. Through mutational and compensatory mutational analysis, we demonstrated this base pairing impairs the inhibitory activity of the U1-binding site. Given the difficulty of prediction of secondary structure, the bioinformatic analysis of such base pairing will be a challenge that we have to meet for future studies.
Lessons for hydrogen bonding between U1 snRNP and the 5′ss
The results of the mutagenesis are also of interest to understand the role of hydrogen bonding in the U1 snRNP-5′ss interaction used during splicing and alternative splicing. There is strong genetic and biochemical evidence that U1 snRNA base pairing to 5′ss is critical for 5′ss selection (7–9). However, there are other factors that contribute to 5′ss choice. For example, SR proteins along with some of the U1 snRNP proteins affect base pairing of U1 snRNA to the 5′ss (35–37). It has been shown that yeast U1C can bind a consensus 5′ss and so it could assist in splice site choice and base pairing of U1 snRNA (35). However, this has only been demonstrated in yeast, where 5′ss sequences are generally consensus and U1 snRNP is more complex than in higher eukaryotes. Also, U5 and U6 snRNPs base pair to the 5′ss and may be preassembled to U1 snRNP before U1 snRNP binding to the 5′ss (38). In spite of this, competition experiments clearly showed that the stability of the duplex formed by U1 snRNA and the 5′ss governs the choice between nearby 5′ss (32). Our results also agree with a strong influence of U1 snRNA binding in 5′ss choice. We believe that the saturation mutagenesis analysis performed in this work allows a good and quantitative method to evaluate cellular U1 snRNA binding to a 5′ss without the confounding affects of U5 or U6 snRNA or other components of the spliceosome. The results reveal that positions 3 to 8 [GGU(A/G)(A/G)G] are critical, but are not sufficient to allow inhibition (Figures 2 and 3). A similar [GGU(A/G)AG] sequence can be found in the Shapiro and Senapathy (39) consensus matrix [(A/C)AGGU(A/G)AGUn], which shows the conservation observed after the alignment of 1446 5′ss sequences. As this matrix should reflect the effect in a 5′ss of several splicing factors, including U5 and U6 snRNPs, we propose that the low conservation of a G at position 7 is not forced by U1 snRNA binding. Similarly, A/G is favored at position 2 in our results whereas it is A in the matrix because splicing requires an interaction between this A and a U from U5 snRNP (40). Nucleotides surrounding the position 3 to 8 nt core seem less critical for U1 snRNA binding. Point mutations are mildly tolerated at position 9, but efficient splicing requires a U to base pair with A from U6 snRNP and this is reflected in the Shapiro and Senapathy consensus matrix. Surprisingly, A or U at position 10 resulted in almost the same level of inhibition, suggesting a similar efficiency of UA or UU base pairing, which is not detected in the rest of the duplex. We cannot exclude that the first A of U1 snRNA or another part of the U1 snRNP complex may interact with the 10U mutant. The fact that the Shapiro and Senapathy matrix does not reflect any preference for position 10 indicates either that A or U nucleotides are detrimental to other factors or that when U1 snRNA binding to the 5′ss is too stable, splicing is less efficient due to a delayed release of U1 snRNP from the spliceosome (40,41).
Noncanonical UU pairings have been described in 5′ss from yeast and human at positions 6 and 7 whereas we find UU pairings are not allowed. Instead, we observe inhibitory activity only with UA standard Watson–Crick base pairings and GU wobble base pairs, both of which contribute to 5′ss selection (32). GU wobble pairs could not only be formed at positions 6 and 7, but also at 1, 2 and 10 (Figure 3). Structural and biochemical studies with model RNA oligonucleotides conclude that a single GU pair within an A helix induces only minor changes in thermodynamic stability consistent with a 2-hydrogen bond pattern of similar but slightly less strength to that of an AU pair. However, the inhibition we observe with all GU containing duplexes is relatively low compared to the wt sequence. This is particularly striking at position 2 where a GU wobble shows reduced activity comparable to a CU and UU mismatch. Also, tandem GU pairs may allow splicing even if they could lead to changes in thermodynamic stability consistent with loss of the 2-hydrogen bond pattern (32,42,43). However, tandem GU wobbles at position 6 and 7 completely abrogate inhibition. The decreased inhibition observed for duplexes with terminal wobbles (positions 1 and 10) is consistent with reports that terminal GU pairs are more unstable than internal GU pairs due to lack of base stacking at the loop side, i.e. the side away from the helix. Importantly, a terminal 5′GU does not equal a terminal 5′UG in that 5′GU is much more thermodynamically stable because of differences in stacking, a phenomena known as the ‘5′-end rule’ (42,44). Both the position 1U and 10G single U1 site mutants produce a terminal 5′U-G pair, the less stable category, a fact consistent with their lowered activity relative to the wtU1 site. As for the significant reduction in activity observed with wobbles at positions 2, 6 and 7 we consider it possible that U1 snRNP proteins can sense a loss in the smooth geometry arising from continuous Watson–Crick pairs. Resolution of these outstanding issues will require a concerted effort involving structural and biochemical approaches.
Models based on ΔG and NH
Our analysis of the duplex formed by the U1 snRNP and the target site indicates that higher inhibition is obtained with duplex lengths of 10–16 nt (Figure 2). For shorter duplexes we find some 8-bp continuous duplexes inhibit while other 8-bp duplexes do not (Figures 2 and 4). Furthermore, the activities of some of the point mutations in Figure 3 were unexpected. In order to develop a model to explain these differences, we took advantage of two recent reports that analyzed an extensive series of point mutations to the canonical 5′ss for changes in splicing efficiency (31,32). Interestingly, both reports proposed distinctly different models to explain the behavior of the 5′ss mutants. In the first case, a model based on free energy (ΔG) calculations of the various U1 snRNA:5′ss duplexes was found to be a good predictor of splicing activity of the 5′ss mutants (32). Thus, an increase in ΔG of the U1 snRNA:5′ss duplex correlated well with an increase in splicing activity. We applied the ΔG model to our mutational analysis of the U1 snRNA:U1 site duplex and found a good correlation with inhibitory activity. However, we also found many exceptions where the correlation broke down, especially for those mutants producing a GU pair that greatly reduced the value of this approach (Gunderson,S.I., unpublished data). Given these exceptions we examined a second published model based on counting the number of continuous hydrogen bonds (NH) in an uninterrupted U1 snRNA:5′ss duplex (31). When NH ≥ 14, this model predicts that the sequence should be active in pre-mRNA splicing. When the helix had mismatches, then NH ≥ 15 was necessary for activity with ≥5 hydrogen bonds occurring 3′ to the mismatch and ≥6 hydrogen bonds 5′ to the mismatch. Notably, this analysis counted GU wobble pairs (but not UU pairs) as 2 hydrogen bonds. In applying this hydrogen bond model to our data, we not only found some correlation but also many inconsistencies. By making modifications of the model, in particular when terminal GU pairs were excluded in the calculation of the NH value, we found a significant improvement in its predictive value. Thus, we observe no activity (<2-fold) for all U1 sites with NH ≤ 18 and good activity (consistently >5-fold) for all U1 sites with NH ≥ 21 (see NH values in Figure 4). The model does not do well, however for U1 sites where NH = 19 or NH = 20 as many sites are inactive whereas others are active (up to 6.8-fold). We tried additional ad hoc modifications to the model but in each case there was little or no improvement. Furthermore, none of the models explain the lowered activity pattern of the 6G and 7G single mutants that make an internal GU pair (see above). Finally, as with any mutational analysis, we cannot formally rule out that a given mutation creates a binding site for a factor that competes with U1 snRNA.
Specificity of U1i
U1 snRNP-mediated inhibition can be detected with some targets of only 8 nt. Thus, U1i could have off-target inhibition levels as seen with RNAi, where 6-nt long targets may function for inhibition. Our results indicate that a 5′-end modified U1 snRNA designed to bind a 10-nt long target, could also interact with sequences with mismatches at the end of the duplex and could also form some GU base pairs. We first thought that binding to mismatches could be driven by high levels of endogenous U1 snRNA (typically 1 million/cell) as this is what was evaluated in the experiments shown in Figures 2–5. However, transfection of a plasmid that expresses a 5′-end mutated U1 snRNA also induces inhibition of the expression of a reporter gene with a target of 8 nt (Figure 6B). We have not yet evaluated whether GU base pairing allows functional inhibition to exogenous U1 snRNAs.
So what are the points in favor of U1i specificity? First, U1i only works in the 3′ terminal exon (1,2). Second, similar to what has been described for U1 snRNA function in splicing (10,33), secondary structures occlude target sequences (Figure 5). Our detailed study of the effect of stem structures concludes that the complete target sequence, that is at least 8 nt, should be accessible to U1 snRNP. Thus, specificity could be greatly enhanced by this fact, as binding sites for U1i must be in the terminal exon of target genes, which are generally enriched in secondary structures. Surprisingly, specificity may also be enhanced because factors that help U1 snRNA binding to a target for splicing actually decrease U1 snRNP-mediated inhibition (Figure 6B).
TIA-1 has been described to increase U1 snRNA binding to weak 5′ss (14). Thus, we rationalized that TIA-1 could help binding of 5′-end mutated U1 snRNAs to sequences that lack perfect complementarity and therefore increase off-target inhibition of U1i. This is important as putative TIA-1-binding sites have been found in the 3′UTR of almost 3% of the genes analyzed, where TIA-1 could function to control mRNA translation (48). Thus, we analyzed the effect on U1i of TIA-1-binding sequences located downstream of a target site in the presence of 5′-end mutated U1 snRNAs that lack perfect complementarity to the target (Figure 6B). Opposite to what we expected, TIA-1-binding sequences abolished U1i when the target had an eight out of 10 complementarity to U1 snRNA (Figure 6B). However, TIA-1 sequences did not affect inhibition of a U1 snRNA with perfect complementarity to the target. This surprising result allows us to suggest that there is a TIA-1-mediated mechanism that increases the specificity of U1i. It will be of interest to determine the molecular mechanisms that allow this effect.
SR proteins bound close to target sequences can also interfere with U1i. Several molecular mechanisms may explain this result. The RS domain of SR proteins has intrinsic affinity for double-stranded RNA regions and can promote U snRNA- pre-mRNA pairing (49). Thus, SR proteins bound close to target sequences could facilitate binding of U1 snRNP in a conformation which is not optimal for mediating U1i effects. We also hypothesized that as SR proteins help U1 snRNP base pairing to the RNA by binding of their RS domain with the RS domain of U1-70K, this could impede interaction of U1-70K with other factors that could be essential for U1i. In agreement with these hypotheses, we show that the RS domain of SR proteins decreases U1i efficiency (Figure 7E). Also, we show that the RS domain of U1-70K is required for U1i (Figure 7C), probably as U1i needs binding of the RS domain of U1-70K to the carboxyl-terminus of poly(A) polymerase, as required for U1 snRNP inhibition of papillomavirus expression. Thus, SR proteins could drive U1 snRNP functionality in a way that a given U1 snRNA:target site complex could only induce commitment to splicing, in the presence of SR proteins, or inhibit polyadenylation, in the absence of SR proteins. U2AF65 RS effects on U1i seem especially complex. Functional U1i is observed when the RS domain of U1-70K is substituted by the RS domain of U2AF65 (Figure 7C) or when this is tethered upstream of a polyadenylation signal (46), while the same RS domain of U2AF65 is able to inhibit U1i when tethered upstream of a target sequence (Figure 7B). Binding of U1 snRNA to the terminal exon should normally induce U1i as few examples exist of pre-mRNA 3′UTRs that bind SR proteins.
The conclusions obtained in this work help us to make some recommendations for effective U1i. As has been previously determined, target sequences should be located in the 3′terminal exon of target genes. Secondary structures should be avoided as well as sequences bound by SR proteins. Target sequences should be avoided that have U-rich sequences downstream. The target should bind nucleotides 1 to 11 or 2 to 11 of the 5′-end modified U1 snRNA. We do not recommend longer sequences as they may force the opening of the first stem of U1 snRNA and they may have a higher risk of off target inhibition. We also recommend avoiding internal G and U nucleotides, as they allow GU wobbles that carry the risk of lowering the specificity. We believe that these recommendations should govern the design of U1i experiments.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Dr Peter G. Stockley and Gabriella Basnak for anti-MS2 antibody. Technical assistance by N. Razquin is thankfully acknowledged. This work was supported by CICYT (SAF2003-01804), CICYT (BIO2006-13225) and through the ‘UTE project CIMA’ to P.F. as well as by NIH grant GM057286 to S.I.G. Funding to pay the Open Access publication charges for this article were provided by the grants listed above.
Conflict of interest statement. None declared.
REFERENCES
- 1.Beckley SA, Liu P, Stover ML, Gunderson SI, Lichtler AC, Rowe DW. Reduction of target gene expression by a modified U1 snRNA. Mol. Cell. Biol. 2001;21:2815–2825. doi: 10.1128/MCB.21.8.2815-2825.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Furth PA, Choe W, Rex JH, Byrne JC, Baker CC. Sequences homologous to 5′ splice sites are required for the inhibitory activity of papillomavirus late 3′ untranslated regions. Mol. Cell. Biol. 1994;14:5278–5289. doi: 10.1128/mcb.14.8.5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fortes P, Cuevas Y, Guan F, Liu P, Pentlicky S, Jung SP, Chantar ML, Prieto J, Rowe D, Gunderson SI. Inhibiting expression of specific genes in mammalian cells using modified U1 snRNPs targeted to terminal exons of pre-mRNA. Proc. Natl Acad. Sci. USA. 2003;100:8264–8269. doi: 10.1073/pnas.1332669100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sajic R, Lee K, Asai K, Sakac D, Branch DR, Upton C, Cochrane A. Use of modified U1 snRNAs to inhibit HIV-1 replication. Nucleic Acids Res. 2007;35:247–255. doi: 10.1093/nar/gkl1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Proudfoot NJ, Furger A, Dye MJ. Integrating mRNA processing with transcription. Cell. 2002;108:501–512. doi: 10.1016/s0092-8674(02)00617-7. [DOI] [PubMed] [Google Scholar]
- 6.Will CL, Luhrmann R. Protein functions in pre-mRNA splicing. Curr. Opin. Cell Biol. 1997;9:320–328. doi: 10.1016/s0955-0674(97)80003-8. [DOI] [PubMed] [Google Scholar]
- 7.Zhuang Y, Weiner AM. A compensatory base change in U1 snRNA suppresses a 5′ splice site mutation. Cell. 1986;46:827–835. doi: 10.1016/0092-8674(86)90064-4. [DOI] [PubMed] [Google Scholar]
- 8.Séraphin B, Kretzner L, Rosbach M. A U1 snRNA:pre-mRNA base pairing interaction is required early in yeast spliceosome assembly but does not uniquely define the 5′splice site. EMBO J. 1988;7:2533–2538. doi: 10.1002/j.1460-2075.1988.tb03101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Siliciano PG, Guthrie C. 5′ splice selection in yeast: genetic alterations in base-pairing with U1 reveal additional requirements. Genes Dev. 1988;2:1258–1267. doi: 10.1101/gad.2.10.1258. [DOI] [PubMed] [Google Scholar]
- 10.Cohen JB, Snow JE, Spencer SD, Levinson AD. Suppression of mammalian 5′splice-site defects by U1 small nuclear RNAs from a distance. Proc. Natl Acad. Sci. USA. 1994;91:10470–10474. doi: 10.1073/pnas.91.22.10470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lo PC, Roy D, Mount SM. Suppressor U1 snRNAs in Drosophila. Genetics. 1994;138:365–378. doi: 10.1093/genetics/138.2.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 13.Lewis JD, Izaurralde E, Jarmolowski A, McGuigan C, Mattaj IW. A nuclear cap-binding complex facilitates association of U1 snRNP with the cap-proximal 5′ splice site. Genes Dev. 1996;10:1683–1698. doi: 10.1101/gad.10.13.1683. [DOI] [PubMed] [Google Scholar]
- 14.Forch P, Puig O, Kedersha N, Martinez C, Granneman S, Seraphin B, Anderson P, Valcarcel J. The apoptosis-promoting factor TIA-1 is a regulator of alternative pre-mRNA splicing. Mol. Cell. 2000;6:1089–1098. doi: 10.1016/s1097-2765(00)00107-6. [DOI] [PubMed] [Google Scholar]
- 15.Förch P, Puig O, Martínez C, Séraphin B, Valcárcel J. The splicing regulator TIA-1 interacts with U1-C to promote U1 snRNP recruitment to 5′ splice sites. EMBO J. 2002;21:6882–6892. doi: 10.1093/emboj/cdf668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu JY, Maniatis,T. Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell. 1993;17:1061–1070. doi: 10.1016/0092-8674(93)90316-i. [DOI] [PubMed] [Google Scholar]
- 17.Goren A, Ram O, Amit M, Keren H, Lev-Maor G, Vig I, Pupko T, Ast G. Comparative analysis identifies exonic splicing regulatory sequences-the complex definition of enhancers and silencers. Mol. Cell. 2006;22:769–781. doi: 10.1016/j.molcel.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 18.Gunderson SI, Polycarpou-Schwarz M, Mattaj IW. U1 snRNP inhibits pre-mRNA polyadenylation through a direct interaction between U170K and poly(A) polymerase. Mol. Cell. 1998;1:255–264. doi: 10.1016/s1097-2765(00)80026-x. [DOI] [PubMed] [Google Scholar]
- 19.Liu P, Kronenberg M, Jiang X, Rowe D. Modified U1 snRNA suppresses expression of a targeted endogenous RNA by inhibiting polyadenylation of the transcript. Nucleic Acids Res. 2004;32:1512–1517. doi: 10.1093/nar/gkh316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dominski Z, Marzluff WF. Formation of the 3′ end of histone mRNA. Gene. 1999;239:1–14. doi: 10.1016/s0378-1119(99)00367-4. [DOI] [PubMed] [Google Scholar]
- 21.Guhaniyogi J, Brewer G. Regulation of mRNA stability in mammalian cells. Gene. 2001;265:11–23. doi: 10.1016/s0378-1119(01)00350-x. [DOI] [PubMed] [Google Scholar]
- 22.Wilusz CJ, Wormington M, Peltz SW. The cap-to-tail guide to mRNA turnover. Nat. Rev. Mol. Cell. Biol. 2001;2:237–246. doi: 10.1038/35067025. [DOI] [PubMed] [Google Scholar]
- 23.Hannon GJ, Rossi JJ. Unlocking the potential of the human genome with RNA interference. Nature. 2004;431:371–378. doi: 10.1038/nature02870. [DOI] [PubMed] [Google Scholar]
- 24.Aparicio O, Razquin N, Zaratiegui M, Narvaiza I, Fortes P. Adenovirus virus-associated RNA is processed to functional interfering RNAs involved in virus production. J. Virol. 2006;80:1376–1384. doi: 10.1128/JVI.80.3.1376-1384.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Narvaiza I, Aparicio O, Vera M, Razquin N, Bortolanza S, Prieto J, Fortes P. Effect of adenovirus-mediated RNA interference on endogenous microRNAs in a mouse model of multidrug resistance protein 2 gene silencing. J. Virol. 2006;80:12236–12247. doi: 10.1128/JVI.01205-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu P, Gucwa A, Stover ML, Buck E, Lichtler A, Rowe D. Analysis of inhibitory action of modified U1 snRNAs on target gene expression: discrimination of two RNA targets differing by a 1 bp mismatch. Nucleic Acids Res. 2002;30:2329–2339. doi: 10.1093/nar/30.11.2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gautheret D, Damberger SH, Gutell RR. G.U base pairing motifs in ribosomal RNA. RNA. 1995;1:807–814. [PMC free article] [PubMed] [Google Scholar]
- 28.Varani G, McClain WH. The G x U wobble base pair. A fundamental building block of RNA structure crucial to RNA function in diverse biological systems. EMBO Rep. 2000;1:18–23. doi: 10.1093/embo-reports/kvd001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hall KB, McLaughlin LW. Properties of a U1/mRNA 5′ splice site duplex containing pseudouridine as measured by thermodynamic and NMR methods. Biochemistry. 1991;30:1795–1801. doi: 10.1021/bi00221a010. [DOI] [PubMed] [Google Scholar]
- 30.Davis DR. Stabilization of RNA stacking by pseudouridine. Nucleic Acid Res. 1995;23:5020–5026. doi: 10.1093/nar/23.24.5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Freund M, Asang C, Kammler S, Konermann C, Krummheuer J, Hipp M, Meyer I, Gierling W, Theiss S, Preuss T, et al. A novel approach to describe a U1 snRNA binding site. Nucleic Acids Res. 2003;31:6963–6975. doi: 10.1093/nar/gkg901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roca X, Sachidanandam R, Krainer AR. Determinants of the inherent strength of human 5′ splice sites. RNA. 2005;11:683–698. doi: 10.1261/rna.2040605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vilardell J, Warner JR. Regulation of splicing at an intermediate step in the formation of the spliceosome. Genes Dev. 1994;8:211–220. doi: 10.1101/gad.8.2.211. [DOI] [PubMed] [Google Scholar]
- 34.Kyriakopoulou C, Larsson P, Liu L, Schuster J, Soderbom F, Kirsebom LA, Virtanen A. U1-like snRNAs lacking complementarity to canonical 5′ splice sites. RNA. 2006;12:1603–1611. doi: 10.1261/rna.26506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Du H, Rosbash M. The U1 snRNP protein U1C recognizes the 5′ splice site in the absence of base pairing. Nature. 2002;419:86–90. doi: 10.1038/nature00947. [DOI] [PubMed] [Google Scholar]
- 36.Kohtz JD, Jamison SF, Will CL, Zuo P, Luhrmann R, Garcia-Blanco MA, Manley JL. Protein-protein interactions and 5′-splice-site recognition in mammalian mRNA precursors. Nature. 1994;368:119–124. doi: 10.1038/368119a0. [DOI] [PubMed] [Google Scholar]
- 37.Chen JY, Stands L, Staley JP, Jackups RR, Latus LJ, Chang TH. Specific alterations of U1-C protein or U1 small nuclear RNA can eliminate the requirement of Prp28p, an essential DEAD box splicing factor. Mol. Cell. 2001;7:227–232. doi: 10.1016/s1097-2765(01)00170-8. [DOI] [PubMed] [Google Scholar]
- 38.Malca H, Shomron N, Ast G. The U1 snRNP base pairs with the 5′ splice site within a penta-snRNP complex. Mol. Cell. Biol. 2003;23:3442–3455. doi: 10.1128/MCB.23.10.3442-3455.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shapiro MB, Senapathy P. RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucleic Acids Res. 1987;15:7155–7174. doi: 10.1093/nar/15.17.7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Staley JP, Guthrie C. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell. 1998;92:315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- 41.Lund M, Kjems J. Defining a 5′ splice site by functional selection in the presence and absence of U1 snRNA 5′ end. RNA. 2002;2:166–179. doi: 10.1017/s1355838202010786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mizuno H, Sundaralingam M. Stacking of Crick Wobble pair and Watson-Crick pair: stability rules of GU pairs at ends of helical stems in tRNAs and the relation to codon-anticodon Wobble interaction. Nucleic Acids Res. 1978;5:4451–4461. doi: 10.1093/nar/5.11.4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen X, McDowell JA, Kierzek R, Krugh TR, Turner DH. Nuclear magnetic resonance spectroscopy and molecular modeling reveal that different hydrogen bonding patterns are possible for G.U pairs: one hydrogen bond for each G.U pair in r(GGCGUGCC)(2) and two for each G.U pair in r(GAGUGCUC)(2) Biochemistry. 2000;39:8970–8982. [PubMed] [Google Scholar]
- 44.Utsunomiya R, Suto K, Balasundaresan D, Fukamizu A, Kumar PK, Mizuno H. Structure of an RNA duplex r(GGCGBrUGCGCU)2 with terminal and internal tandem G.U base pairs. Acta Crystallogr. D Biol. Crystallogr. 2006;62:331–338. doi: 10.1107/S0907444905043210. [DOI] [PubMed] [Google Scholar]
- 45.Guan F, Caratozzolo RM, Goraczniak R, Ho ES, Gunderson SI. A bipartite U1 site represses U1A expression by synergizing with PIE to inhibit nuclear polyadenylation. RNA J. 2007;13:2129–2140. doi: 10.1261/rna.756707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ko B, Gunderson SI. Identification of new poly(A) polymerase-inhibitory proteins capable of regulating pre-mRNA polyadenylation. J. Mol. Biol. 2002;318:1189–1206. doi: 10.1016/s0022-2836(02)00240-1. [DOI] [PubMed] [Google Scholar]
- 47.Vera M, Sobrevals L, Zaratiegui M, Martinez L, Palencia B, Rodríguez CM, Prieto J, Fortes P. Liver transduction with a simian virus 40 vector encoding insulin-like growth factor I reduces hepatic damage and the development of liver cirrhosis. Gene Ther. 2007;14:203–210. doi: 10.1038/sj.gt.3302858. [DOI] [PubMed] [Google Scholar]
- 48.López de Silanes I, Galbán S, Martindale JL, Yang X, Mazan-Mamczarz K, Indig FE, Falco G, Zhan M, Gorospe M. Identification anf functional outcome of mRNAs associated with RNA-binding protein TIA-1. Mol. Cell. Biol. 2005;25:9520–9531. doi: 10.1128/MCB.25.21.9520-9531.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shen H, Green MR. RS domains contact splicing signals and promote splicing by a common mechanism in yeast through humans. Genes Dev. 2006;20:1755–1765. doi: 10.1101/gad.1422106. [DOI] [PMC free article] [PubMed] [Google Scholar]