Abstract
Sequences proximal to transgene integration sites are able to deregulate transgene expression resulting in complex position effect phenotypes. In addition, transgenes integrated as repeated arrays are susceptible to repeat-induced gene silencing. Using a Cre recombinase-based system we have addressed the influence of transgene copy number (CN) on expression of hCD2 transgenes. CN reduction resulted in a decrease, increase or no effect on variegation depending upon the site of integration. This finding argues that repeat-induced gene silencing is not the principle cause of hCD2 transgene variegation. These results also suggest that having more transgene copies can be beneficial at some integration sites. The transgenic lines examined in this report also exhibited a form of imprinting, which was manifested by decreased levels of expression and increased levels of variegation, upon maternal transmission; and this correlated with DNA hypermethylation and a reduction in epigenetic chromatin modifications normally associated with active genes.
INTRODUCTION
Transgenic technology has greatly facilitated our understanding of the mechanisms regulating gene expression (1). However, it became apparent early on that transgene expression profiles did not always faithfully recapitulate that of their endogenous counterparts (2). This has been attributed to a feature of classical transgenesis namely, that transgenes integrate randomly within the genome. As a consequence, sequences proximal to the site of integration can influence transgene expression in both a positive and negative manner. For example, integration of a gene proximal to heterochromatic sequences results in its silencing in a proportion of cells expected to express it (3). Such aberrant expression is termed position effect variegation (PEV) (4,5).
An additional uncontrollable aspect of transgenesis is that transgenes frequently integrate as multiple copies in a concatameric array. It has been observed that some high copy number (CN) transgenic lines suffer from low levels of expression and high levels of variegation (6–12). This has led to the suggestion that the repetitive nature of transgene arrays induces the formation of heterochromatin at the site of integration, thereby driving gene silencing (6,7). Indeed, such repeat-induced gene silencing (RIGS) has been reported in mammals, flies, plants and fungi (6–13).
The propensity of transgenes to PEV and RIGS has led in part to the identification of a growing class of regulatory elements known as locus control regions (LCRs) (14). LCRs are defined by their ability to confer tissue-specific, CN-dependent and position-independent expression upon linked transgenes (14). Thus, LCRs are unique in their ability to protect transgenes against PEV (15). It has been suggested that LCRs exert their effect by ensuring that the transgenic locus adopts a euchromatic conformation in all cells of the expressing lineage, thereby allowing optimal levels of gene expression (16,17). To date, all identified LCRs have been shown to be composed of a number of regulatory elements (enhancers, border elements and/or insulators) all of which cooperate to prevent PEV (15). Deleting one or more of these elements from the LCR results in sensitivity to PEV (17–21).
The hCD2 gene contains an LCR within the 3′ extragenic region, characterized by three DNaseI hypersensitive regions (HSS1-3) (17,18). In mice, hCD2 transgenes carrying the full LCR are resistant to both classical PEV and RIGS, even when present as arrays of more than 50 copies (17). In contrast, transgenes that lack HSS3 of the LCR are often sensitive to PEV regardless of whether they integrate as single or multiple copies (17). In addition, it has been shown that hCD2 transgenes that lack HSS3 and include arrays of CTG or GAA triplet repeats exhibit PEV silencing at all integration sites (22). This suggests that triplet repeats are dominant in inducing gene silencing and heterochromatinization of the transgene array independently of the chromatin state of the locus at the site of integration.
To determine whether or not the hCD2 gene is sensitive to self-induced RIGS, transgenic lines were generated carrying multiple copies of a hCD2 gene with a truncated LCR. Cre recombinase was used to reduce the transgene CN, thus allowing direct comparison of transgenic lines with different CNs, whilst retaining the same integration site. Interestingly, in this experimental system transgene CN reduction resulted in an increase, a decrease or no change in the extent of variegation suggesting that variegation is not a direct result of RIGS, but probably reflects the state of chromatin proximal to the integration site. Taken together, these observations suggest that the relationship between PEV and RIGS is not a simple one.
Our transgenic lines also exhibited an additional aspect of transgenesis that can result in complex transgene expression profiles. Thus, we observed imprinting in four of the six hCD2 transgenic lines reported herein. In these four lines, transgene expression was lower and variegation higher after maternal transmission of the transgene. Maternally transmitted transgenes showed hypermethylation of LCR sequences, and low levels of epigenetic modifications normally associated with active genes. Imprinting in these mice was not affected by CN reduction.
MATERIALS AND METHODS
Cloning
To generate the 1.3loxP vector pBluescript II SK+ was cut with SpeI, blunted [using the DNA Polymerase I Large (Klenow) Fragment] and then cut with BamHI. The VA vector (23) was cut with BamHI–XmnI and the 1.3-kb LCR fragment isolated and ligated into pBluescript II SK+ to generate 1.3LCR-pBS. The 1.3-kb LCR was lifted from the vector by digestion with BamHI–XbaI and ligated into the BamHI–XbaI digested hCD2-Int4 vector to generate hCD2-Int4-1.3. Finally, the hCD2-Int4-1.3 vector was digested with XbaI–NotI and a loxP site generated from annealed oligos ligated in (see below) to give the final construct 1.3loxP. loxP sense oligo:5′CTAGAATAACTTCGTATAATGTATGCTATACGAAGTTATGC3′. loxP anti-sense oligo:5′GGCCGCATAACTTCGTATAGCATACATTATACGAAGTTATT3′
The final vector was linearized by digesting with KpnI–NotI and the hCD2 gene isolated from the plasmid sequences. Isolated fragments were further purified using Elutip columns (Schleicher and Schuell). Pro-nuclei of fertilized oocytes from (CBA/Ca X C57F7BL/10) Fl mice were injected with the purified DNA at a concentration of 1–2 μg/ml TE buffer. Eggs surviving microinjection were transferred into the oviducts of recipient pseudo-pregnant females. Transgenic founders were detected by Southern blot analysis of genomic tail DNA.
Slot blot analysis
Genomic tail DNA (10 μg) was denatured in 400 μl of 0.4 M NaOH for 10 min before neutralization by the addition of 400 μl of 2 M Ammonium acetate. Samples were loaded in duplicate onto a Schleicher and Schuell slot blot apparatus containing a Schleicher and Schuell nitrocellulose membrane pre-wetted in 1 M ammonium acetate. Filters were removed and the DNA fixed by baking at 80°C for 1 h. Hybridizations were carried out essentially as for Southern blot analysis (see below).
McrBC scanning assay and Southern blot analysis
The McrBC assay was adapted from Santoso et al. (24). Fifty micrograms of genomic DNA was digested overnight with the restriction enzyme BamH1. After digestion, DNA was ethanol precipitated and re-suspended in 55 μl of 10 mM Tris (pH 8.0). DNA samples were divided into five 10 μl aliquots and digested as recommended by the manufacturers with increasing amounts (0–40 enzyme units) of McrBC (New England Biolabs) for 35 min at 37°C in 20 μl final volume. The reaction was stopped by the addition of 2 μl of stop solution (100 mM Tris–Cl pH 7.5, 10 mM EDTA pH 7.5, 1% SDS). Samples were resolved on 1% agarose gels containing 1 × TAE and ethidium bromide (5 μg/ml), run at 2 V/cm overnight. After denaturation in 1.5 mM NaCl, 0.5 N NaOH for 40 min and subsequent neutralization in 0.5 M Tris–HCl pH 7.5, 3 M NaCl for 40 min, separated DNA fragments were transferred onto a Hybond-N+ nylon membrane (GE Healthcare), essentially as described in (25). DNA was then fixed onto the membrane by baking at 80°C for 2 h. DNA probes were labelled with [α-32P]dCTP by random priming using Ready-To-Go DNA labelling beads (GE Healthcare) according to the manufacturer's instructions. Unincorporated nucleotides were removed using an illustra sephadex G-50 nick column (GE Healthcare) according to the manufacturer's instructions. The collected probe was denatured by boiling at 100°C for 5 min with 400 μl of denatured salmon sperm (10 mg/ml) before use in hybridization reactions. Membranes were pre-hybridized at 65°C for at least 2 h in hybridization buffer (10 × Denhardts, 3 × SSC, 0.1% SDS, 10% dextran sulphate, 100 μg/ml denatured salmon sperm DNA). Fifty nanograms of labelled denatured probe was added to the hybridization buffer and hybridized overnight at 65°C. Following hybridization, filters were washed in a final stringency of 0.3 × SSC; 0.1% SDS at 65°C.
Flow cytometry
Single cell suspensions were prepared from spleen. For flow cytometry 1 × 106 cells were stained with antibodies in 100 μl ice-cold PBS supplemented with 0.5% BSA and 0.02% sodium azide on ice for 30 min before washing twice with 100 μl ice-cold PBS azide. Cells were re-suspended in 100 μl PBS azide before data acquisition using a FACSCalibur and CELLQUEST software. All antibodies were used at appropriate saturating concentrations. The antibodies used were as follows: hCD2-FITC (RPA-210 eBioscience), CD4-APC (CT-CD4 Caltag) and CD8-PE (CT-CD8 Caltag).
Cell sorting
Single cell suspensions were prepared from spleen and lymph node. Cells were stained at a concentration of 1 × 108 cells/ml in air-buffered IMDM supplemented with 5% FCS with appropriate saturating concentration of antibodies. Cells were incubated with antibodies for 30 min on ice before washing twice with air-buffered IMDM supplemented with 5% FCS. Cells were sorted using a MoFlo cell sorter (Dako). The antibodies used were as follows, hCD2-PE (RPA-210 eBioscience), TCRβ (H57-5097 eBioscience).
Chromatin immuno-precipitation
Sorted populations were re-suspended in warm air-buffered IMDM at a concentration of 1 × 106 cells/ml. Cells were fixed with 1% formaldehyde for 10 min at 37°C. Cross-linking was stopped by the addition of glycine to a final concentration of 0.125 M. Cells were washed twice with ice-cold PBS. Samples were re-suspended in lysis buffer (1%SDS, 10 mM EDTA, 50 mM Tris–HCl pH 8.0) containing protease inhibitors (Roche) at a concentration of 1 × 107 cells/ml and sonicated using a Bioruptor (Diagenode). Sonicated samples were checked on 1% agarose gel and 50 μl sample was reserved as input. Samples were split into aliquots containing 1 × 106 cells, diluted to 1 × 106 cells/ml in dilution buffer (0.1% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris–HCl pH 8.0, 167 mM NaCl) and pre-cleared with protein A/salmon sperm or protein G/salmon sperm agarose beads (Upstate). Samples were incubated overnight with antibodies at 4°C with rotation. Non-specific rabbit IgG was included as a control antibody. Antibody–chromatin complexes were captured with protein A/salmon sperm or protein G/salmon sperm agarose beads for 4 h at 4°C with rotation. Beads were washed once with each of the following buffers: low salt (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris–HCl pH 8.0, 150 mM NaCl), high salt (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris–HCl pH 8.0, 500 mM NaCl), LiCl (0.25 M LiCl, 1% NP-40, 1% sodium deoxycholate, 1 mM EDTA, 10 mM Tris–HCl pH 8.0, 150 mM NaCl) and twice with TE (1 mM EDTA, 10 mM Tris–HCl pH 8.0). DNA–protein complexes were eluted with 1% SDS, 100 mM NaHCO3 and cross-linking reversed by incubating samples at 65°C overnight in the presence of 0.2 M NaCl. Samples were treated with RNase (20 μg/ml), and then with proteinase K (40 μg/ml) for 1 h. DNA was phenol extracted and ethanol precipitated in the presence glycogen (20 ng/ml). Samples were analysed by real time PCR (see below). The following antibodies were used for immuno-precipitation: anti-H3-Tri-meK4 (Abcam ab8580), anti-H3-Tri-meK36 (Abcam ab9050) and anti-acetyl-Histone-H3 (Upstate 06-599), normal rabbit IgG (Upstate 12-370).
Real time PCR
DNA from ChIP samples was diluted 1/10 except for input DNA which was diluted 1/25. Of this dilution, 6 μl was combined with SYBR Green 2 × master mix (applied Bioscience), and 5 pmol of each primer in a total volume of 20 μl. PCR reactions were run on a 7900HT fast real-time PCR system (applied Bioscience) and analysed using SDS 2.2.2 software (applied Bioscience). 1 × (95°C 10 min) 50 × (94°C 30 s. 60°C 30 s. 72°C 40 s). Using CT values obtained from real-time PCR the following equation was used to determine the relative enrichment of each modification: [1/2(I.P.ct − Input.ct)] – [1/2(IgG.ct − Input.ct)]. Primer sequences were as follows.
5′hCD2-F:TGCTCTTCACATGCTCCATG
5′hCD2-R:AAGTATCTGCCAACCCAGAG
Intron4-F:AGGTCATCAGGTAGTCACAG
Intron4-R:CTCTGAAGTGTTCTGCCTTG
5′LCR-F:TAGACCCGTGTCTGCTCATT
5′LCR-R:ATAGGTTGTTGGACCCAGCT
3′LCR-F:AGGTTGCAGTGAGGTGAGAT
3′LCR-R:TAAGGGAATGGCGTATGAGC
G6PD primers have previously been published (26)
G6PD-F:TAACGGAAGTGGGGTCATCC
G6PD-R:TTCGAGATCGCAAAGTCTTGTC
Statistical analysis
For all experiments, statistical analysis was performed in Excel by using an unpaired Student's t-test.
RESULTS
The influence of transgene CN on PEV
Mice carrying large CN transgenes have been reported to show aberrant gene expression and silencing (8,12). As most of the hCD2 transgenic lines previously analysed contain multiple transgene copies, it was important to address the effect of CN on the pattern of hCD2 transgene expression (17,18,22,23,27–30). To this end, a PEV-susceptible hCD2 transgene carrying a truncated 1.3-kb LCR (omitting HSS3) was constructed (1.3loxP). A single loxP site was included at the 3′ of the construct to allow transgene CN reduction following Cre-mediated recombination (Figure 1A). Integration of multiple head-to-tail transgene copies in transgenic mice would theoretically allow Cre-mediated recombination between loxP sites on adjacent copies of the transgene resulting in excision of any number of copies between the loxP sites.
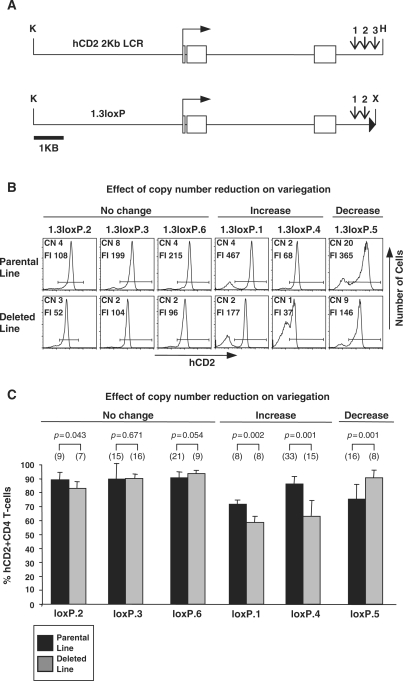
Figure 1.
CN reduction. (A) The hCD2 gene with intron 4 and the 2Kb LCR. Shown below is the 1.3loxP construct containing the truncated 1.3-kb LCR. Open boxes represent exons and filled triangle represents the loxP site. K, KpnI; X, XbaI; H, HinDIII. Vertical arrows indicate positions of DNaseI hypersensitive sites 1–3. (B) Flow cytometric analysis of hCD2 expression on CD4+ T cells from 1.3loxP and 1.3loxPDel transgenic lines. Splenic T cells were isolated, stained with anti-CD4, anti-CD8 and anti-hCD2 and then analysed by flow cytometry. An example FACS plot from each line is shown. CN, transgene CN in each line as determined by slot blot analysis; FI, mean fluorescent intensity of the hCD2+ population (as defined by the horizontal bar). (C) Statistical analysis (unpaired Student's t-test) of variegation in 1.3loxP and 1.3loxPDel transgenic mice. Splenic T cells from 1.3loxP and 1.3loxPDel transgenic mice were analysed by flow cytometry as described above. The percentages of hCD2+ cells are represented. Error bars show standard deviations. Numbers in brackets above the bars indicate the number of mice used from each line. Black bars represent the parental lines, whereas grey bars represent deleted lines. The name of each line has been shortened to from 1.3loxP to loxP.
Following microinjection of the 1.3loxP construct described above, founder mice were bred to CBA non-transgenic mice to establish six independent transgenic lines denoted 1.3loxP.1–6. To mediate transgene CN reduction, each 1.3loxP line was bred to the protamine-Cre (PC3-Cre) transgenic mouse line, in which Cre expression is driven by the protamine 1 gene regulatory elements (31). As the expression of the PC3-Cre transgene is restricted to the haploid spermatids, deletion of the floxed allele occurs only on transgene transmission through the male germline (31). Progeny of 1.3loxP/PC3 double transgenic males, were selected for further breeding to establish new lines with fewer copies of the transgene denoted by the suffix ‘Del’, in which the PC3-Cre transgene had been bred out.
CN analysis of 1.3loxP and 1.3loxPDel transgenic lines
Genomic tail DNA was isolated from 1.3loxP and 1.3loxPDel transgenic mice and subjected to slot blot analysis to determine the approximate transgene CN of each line. Slot blots were probed with the hCD2 cDNA to assess the transgene CN (data not shown). Figure 1B shows the calculated transgene CN in each of the 1.3loxP and 1.3loxPDel transgenic lines. Slot blot analysis revealed that in all of the 1.3loxPDel transgenic lines the CN had been decreased through Cre-mediated recombination. In addition, Southern blot analysis suggested that the original site of integration was maintained (data not shown). Thus, this is an effective system to reduce transgene CNs whilst retaining the same site of integration.
Flow cytometric analysis of peripheral T cells after transgene CN reduction
To assess the affect of transgene CN reduction on hCD2 expression in 1.3loxP transgenic mice, flow cytometry was performed on peripheral T cells from 1.3loxP and 1.3loxPDel transgenic mice. Single cell suspensions from spleen were triple stained with anti-CD4, anti-CD8 and anti-hCD2 before analysis by flow cytometry. Figure 1B shows a representative FACS plot from each line. Figure 1C shows statistical data obtained from analysis of multiple mice (7–33) from each transgenic line.
Comparison of hCD2 expression on 1.3loxP and 1.3loxPDel CD4+ T cells reveals that in each 1.3loxPDel line the reduction in CN is associated with a parallel decrease in the level of expression, as indicated by the mean fluorescent intensity (FI) (Figure 1B). In addition, transgene CN reduction affected the extent of variegation in a number of lines. For example, reducing the CN in the 1.3loxP.1 line from four to two copies resulted in a statistically significant (P = 0.002) decrease in percentage of hCD2 expressing cells from 72% (SD 3) to 58% (SD 3) (Figure 1C). Similarly, reducing the CN in the 1.3loxP.4 line from two to one copy resulted in a statistically significant (P = 0.001) decrease in the percentage of hCD2 expressing cells from 86% (SD 5) to 62% (SD 10). This shows that, in both the 1.3loxP.1 and the 1.3loxP.4 lines, reducing the transgene CN increases transgene variegation.
Interestingly, reducing the transgene CN in the 1.3loxP.5 line from twenty to nine copies resulted in a statistically significant (P = 0.001) increase in the percentage of hCD2 expressing cells from 75% (SD 11) to 91% (SD 4). This shows that, in the 1.3loxP.5 line, reducing the transgene CN decreases transgene variegation.
Finally, reducing the transgene CN in the 1.3loxP.2 (from four to three), 1.3loxP.3 (from eight to two) or 1.3loxP.6 (from four to two) lines did not result in any statistically significant change in the percentage of hCD2 expressing cells.
In summary, deleting copies of the 1.3loxP transgene in vivo affected the pattern of variegation in a manner specific to each transgenic line. Therefore, the relationship between variegation and transgene CN appears to be influenced by the site of integration in the hCD2 transgenic system.
Position effect imprinting in hCD2 transgenic mice
In addition to PEV, it has been shown that a number of transgenes exhibit parent-of-origin dependent expression phenotypes (32–38). To determine whether or not the expression profiles in the 1.3loxP transgenic mice were susceptible to parent-of-origin effects we compared offspring which had inherited the transgenic locus paternally or maternally (Figure 2A). Single cell suspensions from the spleen of mice with either paternally or maternally inherited 1.3loxP transgenes were triple stained with anti-CD4, anti-CD8 and anti-hCD2 before analysis by flow cytometry. Analysis of hCD2 expression in these mice revealed that in four of the six lines the 1.3loxP transgene was subject to parent-of-origin effects or imprinting (Figure 2A). Thus, passage of the transgene through the female germline resulted in offspring, in which expression of the transgene differed, from the expression in those that inherited the transgene through the male germline, in two major ways. Firstly, statistical analysis of hCD2 expression shows that, following maternal transmission, variegation was dramatically increased (Figure 2B) and secondly, that levels of expression of the transgene on hCD2+ cells were reduced (Figure 2C). Moreover, in these four imprinted lines maternal transmission results in extremely variable levels of variegation (as indicated by the large standard deviations in Figure 2B). Notably, the pattern of hCD2 expression is indistinguishable within male and female littermates for all lines, indicating that the variegation profile is not dependent upon the sex of the mouse being analysed, but the parental origin of the transgene (data not shown).
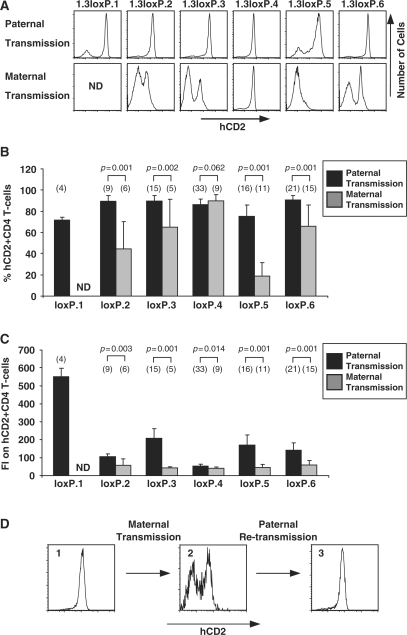
Figure 2.
Imprinted hCD2 transgene expression. (A) Flow cytometric analysis of hCD2 expression on CD4+ T cells from 1.3loxP transgenic lines after paternal or maternal transmission of the transgene. Splenic T cells were isolated, stained with anti-CD4, anti-CD8 and anti-hCD2 and then analysed by flow cytometry. An example FACS plot from each line is shown. ND, not determined. (B) Statistical analysis (unpaired Student's t-test) of variegation in 1.3loxP transgenic mice after paternal or maternal transmission of the transgene. Splenic T cells from 1.3loxP and 1.3loxPDel transgenic mice were analysed by flow cytometry as described above. The percentage of hCD2+ cells is represented. Error bars show standard deviations. Numbers in brackets above the bars indicate the number of mice used from each line. The name of each line has been shortened to from 1.3loxP to loxP. (C) Statistical analysis (unpaired Student's t-test) of the level of hCD2 expression in 1.3loxP transgenic mice after paternal or maternal transmission of the transgene. Splenic T cells from 1.3loxP and 1.3loxPDel transgenic mice were analysed by flow cytometry as described above. The mean fluorescent intensity (FI) of hCD2+ cells is represented. Error bars show standard deviations. Numbers in brackets above the bars indicate the number of mice used from each line. The name of each line has been shortened to from 1.3loxP to loxP. (D) Breeding analysis to show reversal of imprinting. Splenic T cells were isolated, stained with anti-CD4, anti-CD8 and anti-hCD2 and then analysed by flow cytometry. See main text for details.
Importantly, CN reduction did not significantly affect imprinting induced variegation and similarly, the change in variegation resulting from CN reduction, as reported in the previous section (i.e. no change, increase or decrease in variegation) was still evident after maternal transmission of the 1.3loxP and 1.3loxPDel transgenes (data not shown).
The 1.3loxP.1 line was not subjected to imprinting analysis as the transgene is integrated on the Y chromosome (as determined by breeding and FISH analysis; data not shown). Due to imprinting within the 1.3loxP lines Figure 1B and C contain only analysis of 1.3loxP transgenic mice in which the transgene was of paternal origin.
Reversal of imprinting
Most imprinting marks within the genome are reset or erased during gametogenesis (39). Therefore, a breeding programme was established to test whether imprinted expression of the hCD2 transgene obeys the same rules as classical imprinting, i.e. is the imprinting of the transgenic locus reversed after passage through the male germline? Figure 2D shows an example of such an experiment. The first panel displays the characteristic high level of hCD2 expression and low level of variegation after paternal transmission. Maternal transmission from this mouse (Figure 2D panel 1) resulted in the characteristic imprinted expression pattern (Figure 2D panel 2). Subsequent paternal transmission from the mouse in panel 2 resulted in complete reversion to the non-imprinted phenotype (Figure 2D panel 3). The reversal of the imprinted phenotype suggests that imprinting of the 1.3loxP transgene obeys similar rules to classical imprinting.
Imprinted transgenes show DNA hypermethylation
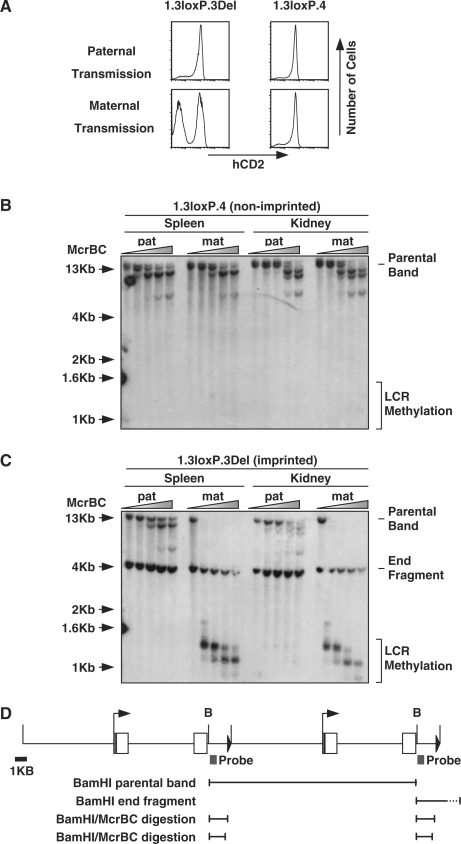
As classical imprinting is associated with DNA methylation we analysed whether imprinted hCD2 transgenes show elevated levels of DNA methylation within the LCR (39). Using an McrBC scanning assay we sought to determine whether the LCR showed differential methylation in non-imprinted and imprinted mice (24). The imprinted line 1.3loxP.3Del was used for methylation analysis, whereas the 1.3loxP.4, which does not display imprinting, was included as a control. These two lines were chosen due to their similar CNs and comparable levels of expression (Figures 1B and 3A).
Figure 3.
Methylation analysis of the hCD2 LCR in expressing and non-expressing tissues. (A) Flow cytometry comparing expression of an imprinted and non-imprinted 1.3loxP transgenic line after paternal and maternal transmission. Splenic T cells were isolated, stained with anti-CD4, anti-CD8 and anti-hCD2 and then analysed by flow cytometry. (B) McrBC analysis of genomic DNA from the non-imprinted 1.3loxP.4 transgenic line. Genomic DNA was extracted from spleen and kidney from 1.3loxP.4 transgenic mice after paternal or maternal transmission of the transgene. Genomic DNA was digested with BamH1 and then incubated with increasing concentrations of McrBC. Treated DNA was analysed by Southern blot using an LCR probe. Shaded triangle represents increasing concentration of McrBC. The position of the parental band and sites of potential LCR methylation are indicated. Mat, maternal transmission; pat, paternal transmission. (C) McrBC analysis of genomic DNA from the imprinted 1.3loxP.3Del transgenic line. Genomic DNA was extracted from spleen and kidney from 1.3loxP.3Del transgenic mice after paternal or maternal transmission of the transgene. Genomic DNA was digested with BamH1 and then incubated with increasing concentrations of McrBC. Treated DNA was then analysed by Southern blot using an LCR probe. Shaded triangle represents increasing concentration of McrBC. The position of the parental band, end fragment and sites of potential LCR methylation are indicated. (D) Diagram of the 1.3loxP transgene and fragments produced in the McrBC assay. Open boxes represent exons and filled triangle represents the loxP site. B, BamHI. Black boxes represent probe used in McrBC assay. Two copies of the transgene are shown integrated in a head-to-tail orientation and shown below are the main fragments resulting from BamH1 or BamHI/McrBC digestion.
Splenic and kidney genomic DNA was extracted from 1.3loxP.3Del and 1.3loxP.4 transgenic mice after either paternal or maternal transmission of the transgene. Extracted genomic DNA was digested with BamHI and then subjected to digestion with increasing amounts of McrBC before analysis by Southern blot using an LCR probe (see Figure 3D for probe details and fragment sizes). In this assay, the appearance of any bands of ≤1.3 kb would indicate the presence of DNA methylation within the hCD2 LCR.
Analysis of genomic DNA extracted from 1.3loxP.4 (non-imprinted) transgenic mice revealed no evidence for LCR-specific DNA methylation in either spleen or kidney after either paternal or maternal transmission of the transgene (Figure 3B). Interestingly, the observed digestion of the 13-kb parent band by McrBC indicates that DNA within the main body of the transgene is methylated in both spleen and kidney (Figure 3B). Furthermore, the level of DNA methylation appears to be marginally higher in the kidney samples. We conclude that in this non-imprinted line the DNA methylation status is not greatly influenced by the parental origin of the transgene.
Analysis of genomic DNA from 1.3loxP.3Del transgenic mice, following paternal transmission, also revealed no evidence for DNA methylation within the hCD2 LCR in either spleen or kidney (Figure 3C). Similar to the analysis of DNA from the non-imprinted line digestion of the 13-kb parent band indicates that DNA within the main body of the transgene is methylated in both spleen and kidney after paternal transmission. In contrast, following maternal transmission of the transgene the presence of bands of <1.3 kb (and the complete disappearance of the 13-kb parent band) provides evidence for high levels of methylation within the hCD2 LCR in both spleen and kidney (Figure 3C).
Differential LCR DNA methylation in hCD2+ and hCD2− T cells from imprinted 1.3loxP.3Del mice
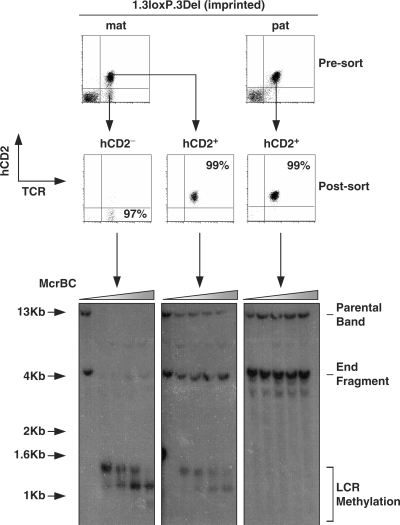
As the previous McrBC assay was performed on single cell suspension from whole organs, it did not allow us to discriminate between T cells that did or did not express the transgene. Thus, we extended the studies to determine whether the methylation status of the transgene is different between expressing and non-expressing lymphocytes. To this end, hCD2+ and hCD2− T cells were FACS sorted from spleen and lymph node of 1.3loxP.3Del mice, after maternal or paternal transmission (Figure 4). DNA extracted from the sorted populations was subjected to McrBC analysis as described in Figure 3. In agreement with data presented above, hCD2+ T cells showed no methylation of the LCR after paternal transmission of the transgene. In contrast, hCD2− cells showed a high degree of DNA methylation after maternal transmission. Interestingly, hCD2+ T cells from the same animals showed a small degree of DNA methylation.
Figure 4.
Methylation analysis in sorted T-cell populations. A McrBC analysis of sorted T-cell populations from 1.3loxP.3Del transgenic mice. Spleen- and lymph node-derived T cells were stained with anti-TCR and anti-hCD2 before flow sorting. T cells (TCR+) were sorted into populations that were either hCD2+ or hCD2−. Each population was at least 97% pure. Genomic DNA was extracted from the sorted populations, digested with BamH1 and then incubated with increasing concentrations of McrBC. Treated DNA was analysed by Southern blot using an LCR probe. Shaded triangle represents increasing concentration of McrBC. The position of the parental band, end fragment and sites of potential LCR methylation are indicated.
ChIP analysis of the hCD2 gene in hCD2+ and hCD2− T cells from imprinted 1.3loxP.3Del mice
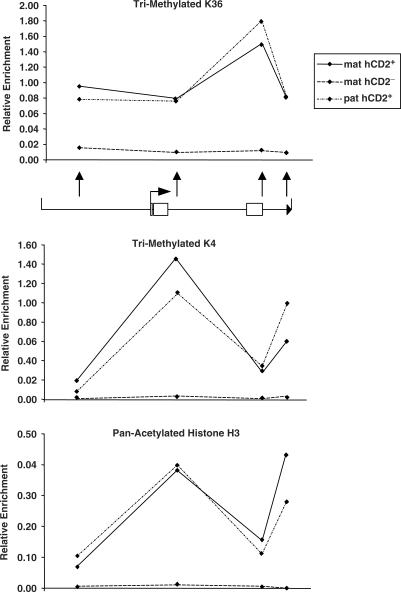
In order to assess the pattern of epigenetic modifications involved in gene regulation ChIP analysis was performed on the sorted populations described above (Figure 5). Analysis of H3-Me3K36 levels, a marker for active genes, revealed enrichment along the hCD2 gene in hCD2+ cells after maternal and paternal transmission, when compared to hCD2− cells. Interestingly, the level of H3-Me3K36 was highest at the 5′ end of the LCR. Analysis of H3-Me3K4 levels revealed a slightly different finding. Although hCD2+ cells from both maternal and paternal transmission showed enrichment for this modification, the levels were highest within hCD2 intron 4 and the 3′ of the LCR which contains the hCD2 enhancer (Figure 5). Analysis of pan-acetylated histone H3 levels revealed a similar profile with enrichment at intron 4 and the 3′ of the LCR in hCD2+ cells after paternal or maternal transmission (Figure 5). All three modifications were absent from the chromatin of the transgenic locus in cells that do not express the transgene.
Figure 5.
ChIP analysis of sorted T-cell populations. ChIP analysis of the hCD2 gene in sorted populations from 1.3loxP3Del transgenic mice. Sorted populations of hCD2+ and hCD2− cells (as described in Figure 4) were fixed in formaldehyde and then subjected to ChIP analysis. Graphs show levels of H3-Tri-meK36, H3-Tri-meK4 and pan-acetyl-Histone-H3 along the hCD2 gene as determined by real-time PCR. Each immuno-precipitation represents average values of two independent experiments and each PCR was repeated in quadruplicate. See ‘Materials and methods’ section for normalization methods. Below the upper graph is a schematic of the transgene and black arrows indicate positions of ChIP primers.
DISCUSSION
Transgene CN and variegation
Sequences proximal to transgene integration sites are able to deregulate transgene expression resulting in complex position effect phenotypes (40). In addition, transgenes integrated as repeated arrays are susceptible to RIGS (41). It has been proposed that RIGS has evolved as a mechanism to silence parasitic repeats within the genome (41). RIGS also may serve a structural role, by providing a sequence-independent mechanism to maintain centromeric integrity through the heterochromatinization of centromere proximal repeats (41).
Previously, nucleotide triplet-repeat expansion has been shown to drive hCD2 transgene silencing in a position-independent manner (22). However, it was not clear whether concatamers of an hCD2 transgene are susceptible to self-induced RIGS. It was therefore interesting to directly address the influence of transgene CN on expression of variegating hCD2 transgenes.
Six hCD2 transgenic lines with a truncated LCR reported herein displayed variegation. Utilizing a Cre recombinase-based approach we were able to reduce the transgene CN in each of the hCD2 transgenic lines. In each of the transgenic lines CN reduction was associated with a concomitant decrease in the level of hCD2 expression on T cells. In addition, CN reduction altered the extent of variegation in a manner that was specific to each line, suggesting that RIGS is not the direct causative agent of variegation in hCD2 transgenic mice, instead the sequences proximal to the site of integration appear to be responsible for the transgene deregulation. Indeed, previous observations from our laboratory, have revealed that not all high-CN hCD2 transgenic lines display variegation even though they possess a disabled/truncated LCR, providing additional evidence to support this hypothesis (17). These results are in contrast with data obtained with triplet-repeat expansion induced RIGS of hCD2 transgenes, which occurs at all integration sites regardless of transgene CN (22). In this system, deregulation appears to be defined by the triplet-repeat itself and is dominant over the influence of sequences proximal to the site of integration.
The observation that variegation increases upon transgene CN reduction indicates that at some integration sites larger transgene arrays have an advantage over smaller arrays, suggesting that having more transgene copies within an array can actually be beneficial rather than a cause of deregulation. It is possible that transgenes embedded within the centre of a large repeat may be shielded better from the influence of neighbouring sequences at the site of integration than when present in a small repeat. Alternatively, regulatory elements on neighbouring transgenes within the array may co-operate in cis to overcome position effects. Such a mechanism has previously been described in λ5-VpreB1 transgenic mice (42).
Our data contrast with results from a number of published studies. For example, Garrick et al. (8) utilized a Cre recombinase-based strategy to reduce CN of an α globin-lacZ transgene, whilst retaining the same integration site. They reported a strict correlation with high transgene CNs and gene silencing. In that study, Garrick et al. utilized transgenic lines with CNs in excess of 100, thus, the relatively low CN of the transgene arrays reported by us herein may be too small to induce true repeat-induced gene silencing. In agreement with this possibility is the observation that the only line to display a decrease in variegation on CN reduction also had the highest starting CN (20 copies). However, it has been reported that in some cases <20 copies are sufficient to drive RIGS in mammalian cells in tissue culture, and CNs as low as three are sufficient to drive RIGS in Drosophila and Arabidopsis (7,9,10,13).
Perhaps more importantly, the data reported herein utilized a mammalian promoter and a mammalian reporter gene, whilst a number of the systems previously described rely upon prokaryotic reporter genes. It is worth noting that genes of prokaryotic origin are exceptionally susceptible to PEV (43).
At first glance, these disparate results suggest that different mechanisms of gene silencing are employed in each system. However, it is likely that these differences simply reflect the presence of a broad spectrum of gene/sequence-specific sensitivity to RIGS. For example, in Drosophila, variegation of the white gene is sensitive to RIGS at all chromosomal locations, whereas, RIGS of the brown gene is absolutely dependent upon the proximity to heterochromatic sequences (7,10). Thus, the composition of the repeat is important in determining sensitivity to RIGS.
Imprinted position effect variegation
Flow cytometry revealed that in a number of 1.3loxP lines transgene expression was sensitive to the parental origin of the transgene. In four of the 1.3loxP lines, maternal transmission of the transgene resulted in both an increase in variegation and a decrease in the level of transgene expression on hCD2+ cells.
Imprinting was reversed or erased on subsequent passage through the male germline. This characteristic, normally associated with ‘classical imprinting’, suggests that similar mechanisms may be involved in imprinting of the 1.3loxP transgenes.
As hCD2 transgenes with the full LCR do not show imprinted expression, it is unlikely that this finding represents a normal mode of hCD2 regulation. Currently it remains unclear as to why the 1.3loxP transgenes should be so susceptible to parent-of-origin effects. One possible explanation is that the hCD2 transgenes in the 1.3loxP mice could be integrated proximal to naturally imprinted genes. In addition, a combination of genome-wide and chromosome-specific analysis of CpG methylation sites has identified genomic sites that do not contain imprinted genes but display monoallelic parent-of-origin specific DNA methylation (44,45). Thus, it is possible that imprinting has resulted from transgene integration proximal to such an area of differential methylation. We hypothesize that all hCD2 transgenes integrated at such sites would become methylated on maternal transmission. However, in the absence of HSS3 the LCR would remain methylated in T cells, resulting in its deregulation.
In support of this hypothesis, we have shown using the McrBC scanning assay that in imprinted lines the LCR is highly methylated in both expressing and non-expressing tissues after maternal transmission of the transgene. This suggests that DNA methylation may be involved in imprinted transgene silencing. However, McrBC analysis on sorted populations of T cells revealed that even in hCD2 expressing cells the LCR is methylated to some extent after maternal transmission of the imprinted transgene. This indicates that complete removal of DNA methylation from all copies of the transgene is not absolutely essential for transgene expression. However, as this analysis was performed on a bulk population of cells we do not know if this intermediate level of methylation represents a fraction of cells carrying high levels of methylation (possibly indicative of cells that have recently escaped silencing/or about to be silenced) or whether all cells within the population carry an intermediate level of methylation. Alternatively, this intermediate level could arise if some of the transgene copies within the array are methylated.
It is interesting to note that the methylated sequences within the hCD2 LCR in imprinted mice correlate approximately with the position of an Alu repeat. Alu repeats are frequently methylated within the genome, perhaps to silence these otherwise transposable elements. Furthermore, it appears that some endogenous Alu repeats are subject to parent-of-origin differential methylation (46,47).
ChIP analysis revealed that the hCD2− population showed lower levels of epigenetic modifications normally associated with active genes (H3-Me3K36, H3-Me3K4 and acetylation of histone H3). Interestingly, levels of H3-Me3K36 were highest at the 5′ end of the LCR. This fits with previously published data in which H3-Me3K36 is enriched in the 3′ of transcribed genes (48,49). In addition, levels of H3-Me3K4 and acetylation of histone H3 were enriched within intron 4 and the 3′ of the LCR, both of which contain hypersensitive sites. This is in agreement with previously published data describing association of these modifications with DNaseI hypersensitive sites and regulatory elements (50,51).
As mentioned previously imprinted transgene expression has been described in a number of systems (32–38). In addition, imprinted transgenes consistently show lower levels of expression and DNA hypermethylation after maternal transmission (32–38). In our system, we propose a mechanism where imprinted position effects result in parental-specific methylation of LCR sequences. This methylation in turn prevents acquisition of appropriate histone modifications along the hCD2 gene in developing T cells, resulting in failure to activate the transgene, thereby driving imprinted variegation. Although the mechanisms for transgene imprinting have yet to be resolved, the similarity of these previously published results and the results reported herein suggest that related imprinting mechanisms are involved in each case. Determining why these transgenes are susceptible to imprinting and the identification of factors important in directing the process could reveal invaluable information regarding the mechanisms of classical imprinting.
ACKNOWLEDGEMENTS
We wish to thank Aaron Rae and Graham Preece for cell sorting. This work was supported by the MRC. H V-F was supported by European Union (LSHG-CT-2003-503259). Funding to pay the Open Access publication charges for this article was provided by European Commission, Sixth Research and Technological Development Framework Programme.
Conflict of interest statement. None declared.
REFERENCES
- 1.Babinet C, Morello D, Renard JP. Transgenic mice. Genome. 1989;31:938–949. doi: 10.1139/g89-165. [DOI] [PubMed] [Google Scholar]
- 2.Pinkert CA, Widera G, Cowing C, Heber-Katz E, Palmiter RD, Flavell RA, Brinster RL. Tissue-specific, inducible and functional expression of the E alpha d MHC class II gene in transgenic mice. EMBO J. 1985;4:2225–2230. doi: 10.1002/j.1460-2075.1985.tb03918.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Muller HJ. Types of visable variations induced by X-rays in Drosophila. J. Genet. 1930;22:299–334. [Google Scholar]
- 4.Allshire RC, Javerzat JP, Redhead NJ, Cranston G. Position effect variegation at fission yeast centromeres. Cell. 1994;76:157–169. doi: 10.1016/0092-8674(94)90180-5. [DOI] [PubMed] [Google Scholar]
- 5.Eissenberg JC. Position effect variegation in Drosophila: towards a genetics of chromatin assembly. Bioessays. 1989;11:14–17. doi: 10.1002/bies.950110105. [DOI] [PubMed] [Google Scholar]
- 6.Assaad FF, Tucker KL, Signer ER. Epigenetic repeat-induced gene silencing (RIGS) in Arabidopsis. Plant Mol. Biol. 1993;22:1067–1085. doi: 10.1007/BF00028978. [DOI] [PubMed] [Google Scholar]
- 7.Dorer DR, Henikoff S. Expansions of transgene repeats cause heterochromatin formation and gene silencing in Drosophila. Cell. 1994;77:993–1002. doi: 10.1016/0092-8674(94)90439-1. [DOI] [PubMed] [Google Scholar]
- 8.Garrick D, Fiering S, Martin DI, Whitelaw E. Repeat-induced gene silencing in mammals. Nat. Genet. 1998;18:56–59. doi: 10.1038/ng0198-56. [DOI] [PubMed] [Google Scholar]
- 9.McBurney MW, Mai T, Yang X, Jardine K. Evidence for repeat-induced gene silencing in cultured Mammalian cells: inactivation of tandem repeats of transfected genes. Exp. Cell Res. 2002;274:1–8. doi: 10.1006/excr.2001.5443. [DOI] [PubMed] [Google Scholar]
- 10.Sabl JF, Henikoff S. Copy number and orientation determine the susceptibility of a gene to silencing by nearby heterochromatin in Drosophila. Genetics. 1996;142:447–458. doi: 10.1093/genetics/142.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ye F, Signer ER. RIGS (repeat-induced gene silencing) in Arabidopsis is transcriptional and alters chromatin configuration. Proc. Natl Acad. Sci. USA. 1996;93:10881–10886. doi: 10.1073/pnas.93.20.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davis BP, MacDonald RJ. Limited transcription of rat elastase I transgene repeats in transgenic mice. Genes Dev. 1988;2:13–22. doi: 10.1101/gad.2.1.13. [DOI] [PubMed] [Google Scholar]
- 13.Barry C, Faugeron G, Rossignol JL. Methylation induced premeiotically in Ascobolus: coextension with DNA repeat lengths and effect on transcript elongation. Proc. Natl Acad. Sci. USA. 1993;90:4557–4561. doi: 10.1073/pnas.90.10.4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grosveld F, van Assendelft GB, Greaves DR, Kollias G. Position-independent, high-level expression of the human beta-globin gene in transgenic mice. Cell. 1987;51:975–985. doi: 10.1016/0092-8674(87)90584-8. [DOI] [PubMed] [Google Scholar]
- 15.Li Q, Harju S, Peterson KR. Locus control regions: coming of age at a decade plus. Trends Genet. 1999;15:403–408. doi: 10.1016/s0168-9525(99)01780-1. [DOI] [PubMed] [Google Scholar]
- 16.Kioussis D, Vanin E, deLange T, Flavell RA, Grosveld FG. Beta-globin gene inactivation by DNA translocation in gamma beta-thalassaemia. Nature. 1983;306:662–666. doi: 10.1038/306662a0. [DOI] [PubMed] [Google Scholar]
- 17.Festenstein R, Tolaini M, Corbella P, Mamalaki C, Parrington J, Fox M, Miliou A, Jones M, Kioussis D. Locus control region function and heterochromatin-induced position effect variegation. Science. 1996;271:1123–1125. doi: 10.1126/science.271.5252.1123. [DOI] [PubMed] [Google Scholar]
- 18.Lang G, Mamalaki C, Greenberg D, Yannoutsos N, Kioussis D. Deletion analysis of the human CD2 gene locus control region in transgenic mice. Nucleic Acids Res. 1991;19:5851–5856. doi: 10.1093/nar/19.21.5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Caterina JJ, Ryan TM, Pawlik KM, Palmiter RD, Brinster RL, Behringer RR, Townes TM. Human beta-globin locus control region: analysis of the 5′ DNase I hypersensitive site HS 2 in transgenic mice. Proc. Natl Acad. Sci. USA. 1991;88:1626–1630. doi: 10.1073/pnas.88.5.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee GR, Fields PE, Griffin TJ, Flavell RA. Regulation of the Th2 cytokine locus by a locus control region. Immunity. 2003;19:145–153. doi: 10.1016/s1074-7613(03)00179-1. [DOI] [PubMed] [Google Scholar]
- 21.Ronai D, Berru M, Shulman MJ. Variegated expression of the endogenous immunoglobulin heavy-chain gene in the absence of the intronic locus control region. Mol. Cell. Biol. 1999;19:7031–7040. doi: 10.1128/mcb.19.10.7031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saveliev A, Everett C, Sharpe T, Webster Z, Festenstein R. DNA triplet repeats mediate heterochromatin-protein-1-sensitive variegated gene silencing. Nature. 2003;422:909–913. doi: 10.1038/nature01596. [DOI] [PubMed] [Google Scholar]
- 23.Zhumabekov T, Corbella P, Tolaini M, Kioussis D. Improved version of a human CD2 minigene based vector for T cell-specific expression in transgenic mice. J. Immunol. Methods. 1995;185:133–140. doi: 10.1016/0022-1759(95)00124-s. [DOI] [PubMed] [Google Scholar]
- 24.Santoso B, Ortiz BD, Winoto A. Control of organ-specific demethylation by an element of the T-cell receptor-alpha locus control region. J. Biol. Chem. 2000;275:1952–1958. doi: 10.1074/jbc.275.3.1952. [DOI] [PubMed] [Google Scholar]
- 25.Southern EM. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 26.Harker N, Naito T, Cortes M, Hostert A, Hirschberg S, Tolaini M, Roderick K, Georgopoulos K, Kioussis D. The CD8alpha gene locus is regulated by the Ikaros family of proteins. Mol. Cell. 2002;10:1403–1415. doi: 10.1016/s1097-2765(02)00711-6. [DOI] [PubMed] [Google Scholar]
- 27.Lang G, Wotton D, Owen MJ, Sewell WA, Brown MH, Mason DY, Crumpton MJ, Kioussis D. The structure of the human CD2 gene and its expression in transgenic mice. EMBO J. 1988;7:1675–1682. doi: 10.1002/j.1460-2075.1988.tb02995.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Greaves DR, Wilson FD, Lang G, Kioussis D. Human CD2 3′-flanking sequences confer high-level, T cell-specific, position-independent gene expression in transgenic mice. Cell. 1989;56:979–986. doi: 10.1016/0092-8674(89)90631-4. [DOI] [PubMed] [Google Scholar]
- 29.Lake RA, Wotton D, Owen MJ. A 3′ transcriptional enhancer regulates tissue-specific expression of the human CD2 gene. EMBO J. 1990;9:3129–3136. doi: 10.1002/j.1460-2075.1990.tb07510.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wotton D, Flanagan BF, Owen MJ. Chromatin configuration of the human CD2 gene locus during T-cell development. Proc. Natl Acad. Sci. USA. 1989;86:4195–4199. doi: 10.1073/pnas.86.11.4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O’Gorman S, Dagenais NA, Qian M, Marchuk Y. Protamine-Cre recombinase transgenes efficiently recombine target sequences in the male germ line of mice, but not in embryonic stem cells. Proc. Natl Acad. Sci. USA. 1997;94:14602–14607. doi: 10.1073/pnas.94.26.14602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chaillet JR, Bader DS, Leder P. Regulation of genomic imprinting by gametic and embryonic processes. Genes Dev. 1995;9:1177–1187. doi: 10.1101/gad.9.10.1177. [DOI] [PubMed] [Google Scholar]
- 33.Chaillet JR, Vogt TF, Beier DR, Leder P. Parental-specific methylation of an imprinted transgene is established during gametogenesis and progressively changes during embryogenesis. Cell. 1991;66:77–83. doi: 10.1016/0092-8674(91)90140-t. [DOI] [PubMed] [Google Scholar]
- 34.Kearns M, Preis J, McDonald M, Morris C, Whitelaw E. Complex patterns of inheritance of an imprinted murine transgene suggest incomplete germline erasure. Nucleic Acids Res. 2000;28:3301–3309. doi: 10.1093/nar/28.17.3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Preis JI, Downes M, Oates NA, Rasko JE, Whitelaw E. Sensitive flow cytometric analysis reveals a novel type of parent-of-origin effect in the mouse genome. Curr. Biol. 2003;13:955–959. doi: 10.1016/s0960-9822(03)00335-x. [DOI] [PubMed] [Google Scholar]
- 36.Reik W, Collick A, Norris ML, Barton SC, Surani MA. Genomic imprinting determines methylation of parental alleles in transgenic mice. Nature. 1987;328:248–251. doi: 10.1038/328248a0. [DOI] [PubMed] [Google Scholar]
- 37.Ueda T, Yamazaki K, Suzuki R, Fujimoto H, Sasaki H, Sakaki Y, Higashinakagawa T. Parental methylation patterns of a transgenic locus in adult somatic tissues are imprinted during gametogenesis. Development. 1992;116:831–839. doi: 10.1242/dev.116.4.831. [DOI] [PubMed] [Google Scholar]
- 38.Swain JL, Stewart TA, Leder P. Parental legacy determines methylation and expression of an autosomal transgene: a molecular mechanism for parental imprinting. Cell. 1987;50:719–727. doi: 10.1016/0092-8674(87)90330-8. [DOI] [PubMed] [Google Scholar]
- 39.da Rocha ST, Ferguson-Smith AC. Genomic imprinting. Curr. Biol. 2004;14:R646–R649. doi: 10.1016/j.cub.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 40.Clark AJ, Bissinger P, Bullock DW, Damak S, Wallace R, Whitelaw CB, Yull F. Chromosomal position effects and the modulation of transgene expression. Reprod. Fertil. Dev. 1994;6:589–598. doi: 10.1071/rd9940589. [DOI] [PubMed] [Google Scholar]
- 41.Henikoff S. Conspiracy of silence among repeated transgenes. Bioessays. 1998;20:532–535. doi: 10.1002/(SICI)1521-1878(199807)20:7<532::AID-BIES3>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 42.Sabbattini P, Georgiou A, Sinclair C, Dillon N. Analysis of mice with single and multiple copies of transgenes reveals a novel arrangement for the lambda5-VpreB1 locus control region. Mol. Cell Biol. 1999;19:671–679. doi: 10.1128/mcb.19.1.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Montoliu L, Chavez S, Vidal M. Variegation associated with lacZ in transgenic animals: a warning note. Transgenic Res. 2000;9:237–239. doi: 10.1023/a:1008995730285. [DOI] [PubMed] [Google Scholar]
- 44.Yamada Y, Watanabe H, Miura F, Soejima H, Uchiyama M, Iwasaka T, Mukai T, Sakaki Y, Ito T. A comprehensive analysis of allelic methylation status of CpG islands on human chromosome 21q. Genome Res. 2004;14:247–266. doi: 10.1101/gr.1351604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Strichman-Almashanu LZ, Lee RS, Onyango PO, Perlman E, Flam F, Frieman MB, Feinberg AP. A genome-wide screen for normally methylated human CpG islands that can identify novel imprinted genes. Genome Res. 2002;12:543–554. doi: 10.1101/gr.224102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hellmann-Blumberg U, Hintz MF, Gatewood JM, Schmid CW. Developmental differences in methylation of human Alu repeats. Mol. Cell Biol. 1993;13:4523–4530. doi: 10.1128/mcb.13.8.4523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sandovici I, Kassovska-Bratinova S, Loredo-Osti JC, Leppert M, Suarez A, Stewart R, Bautista FD, Schiraldi M, Sapienza C. Interindividual variability and parent of origin DNA methylation differences at specific human Alu elements. Hum. Mol. Genet. 2005;14:2135–2143. doi: 10.1093/hmg/ddi218. [DOI] [PubMed] [Google Scholar]
- 48.Bannister AJ, Schneider R, Myers FA, Thorne AW, Crane-Robinson C, Kouzarides T. Spatial distribution of di- and tri-methyl lysine 36 of histone H3 at active genes. J. Biol. Chem. 2005;280:17732–17736. doi: 10.1074/jbc.M500796200. [DOI] [PubMed] [Google Scholar]
- 49.Farris SD, Rubio ED, Moon JJ, Gombert WM, Nelson BH, Krumm A. Transcription-induced chromatin remodeling at the c-myc gene involves the local exchange of histone H2A.Z. J. Biol. Chem. 2005;280:25298–25303. doi: 10.1074/jbc.M501784200. [DOI] [PubMed] [Google Scholar]
- 50.Bulger M, Schubeler D, Bender MA, Hamilton J, Farrell CM, Hardison RC, Groudine M. A complex chromatin landscape revealed by patterns of nuclease sensitivity and histone modification within the mouse beta-globin locus. Mol. Cell Biol. 2003;23:5234–5244. doi: 10.1128/MCB.23.15.5234-5244.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang H, Yan C, Asangani I, Allgayer H, Boyd DD. Identification of an histone H3 acetylated/K4-methylated-bound intragenic enhancer regulatory for urokinase receptor expression. Oncogene. 2007;26:2058–2070. doi: 10.1038/sj.onc.1210003. [DOI] [PubMed] [Google Scholar]