Abstract
The rate of DNA supercoil removal by human topoisomerase IB (TopIB) is slowed down by the presence of the camptothecin class of antitumor drugs. By preventing religation, these drugs also prolong the lifetime of the covalent TopIB–DNA complex. Here, we use magnetic tweezers to measure the rate of supercoil removal by drug-bound TopIB at a single DNA sequence in real time. This is accomplished by covalently linking camptothecins to a triple helix-forming oligonucleotide that binds at one location on the DNA molecule monitored. Surprisingly, we find that the DNA dynamics with the TopIB–drug interaction restricted to a single DNA sequence are indistinguishable from the dynamics observed when the TopIB–drug interaction takes place at multiple sites. Specifically, the DNA sequence does not affect the instantaneous supercoil removal rate or the degree to which camptothecins increase the lifetime of the covalent complex. Our data suggest that sequence-dependent dynamics need not to be taken into account in efforts to develop novel camptothecins.
INTRODUCTION
Eukaryotic Topoisomerase I (TopIB) is an enzyme that allows for the removal of torsional stress from double-stranded (ds)DNA molecules (1–4). It has been proposed that TopIB acts in vivo as a swivelase ahead of a replication fork to remove positive supercoils (5). TopIB functions as a monomer and circumscribes the DNA duplex (6,7), after which the active-site tyrosine attacks the scissile phosphodiester of one of the strands of the DNA duplex, generating a DNA-(3′-phosphotyrosyl)-enzyme intermediate, as well as a free 5′-OH DNA end. The torsional energy that is stored in supercoiled DNA will then power rotation of the 5′-OH end about the intact strand and consequently, supercoils are removed through a swivel mechanism that is termed ‘hindered’, in reference to the significant interactions between the spinning DNA and the enzyme's central cavity (8–10). Religation of the DNA, which arrests the swivelling and thus terminates supercoil removal, appears to be a stochastic process, as the number of supercoils that are removed from the DNA prior to religation follows an exponential distribution (8).
Although the transient covalent DNA-enzyme intermediate nullifies the requirement for an external energy source fueling supercoil removal, the establishment of the DNA-linked TopIB does constitute a risky catalytic route that would pose a danger to the cell if the religation reaction were prohibited. Small planar molecules, such as the camptothecin class of antitumor drugs, can inhibit religation and indeed are cytotoxic. They do so by entering the nick that is created by TopIB and mimicking a DNA base pair, locally deforming the DNA duplex in the process (11–16). Binding of the drug results in a vast decrease in the rate of religation and thus in an increase in the time the covalent complex remains trapped on the DNA (17–21). The generally held paradigm for drug-induced cytotoxicity is that the advancing replication machinery ‘collides’ with the trapped TopIB–CPT–DNA ternary complex, giving rise to lethal DNA lesions and fork collapse (12,14,22,23). However, a combination of single-molecule and yeast-based experiments has recently identified an additional manifestation of drug-induced cytotoxicity, namely that upon treatment with camptothecin, positive supercoils accumulate in a cell cycle-independent manner, presumably owing to a preferential slow-down of drug-mediated positive supercoil relaxation (19).
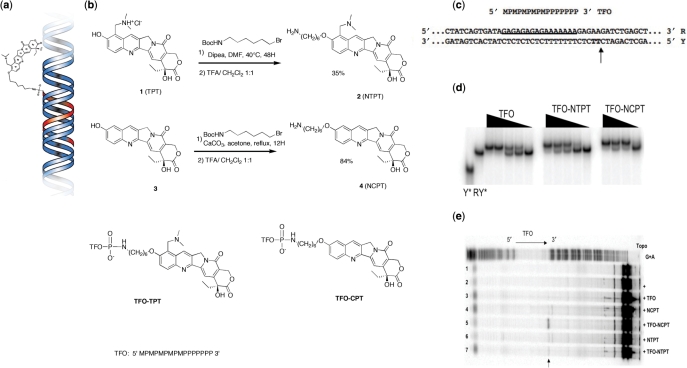
The DNA sequence at which cleavage by human TopIB occurs is only mildly specific (2) and the drug-induced stabilization of the covalent complex occurs at a subset of these sites (12,24). Still, for all practical purposes, the cleavage by TopIB can be considered independent of DNA sequence (25). Indeed, drug treatment leads to widespread DNA damage in cells that affects a multitude of genes, whether specific to cancer cells or not (14,24). One strategy to improve the overall efficacy of the camptothecins is to render them DNA sequence-specific, so as to inflict damage onto oncogenes in cancer cells (26). An implementation of this strategy is the use of triple helix-forming oligonucleotides (TFOs) conjugated to CPT or topotecan (TPT) (27,28). A TPT molecule conjugated to a triple helix bound in the DNA duplex is schematically shown in Figure 1a. These constructs have previously been shown to direct the drug-stabilized covalent complex to specific sequences both in vitro and in cells (29).
Figure 1.
Binding of the TFO–TPT construct and site-specific cleavage by TopIB. (a) A triple helix-forming oligonucleotide (red), covalently linked to a topotecan (TPT) molecule binds in the major groove of DNA (blue). (b) Synthetic route for the preparation of 10-(6-aminohexyloxy)-topotecan NTPT (referred to as TPT) and 10-(6-aminohexyloxy)-camptothecin NCPT (referred to as CPT). The chemical structure of the conjugates TFO–TPT and TFO–CPT. (c) The target site on the duplex is underlined and the topoisomerase I-mediated DNA cleavage site is indicated by an arrow. M, 5-methyl-2′-deoxycytidine; P, 5-propynyl-2′-deoxyuridine. (d) PAGE analysis of triple helix formation: the duplex target RY* ([RY*] = 20 nM), radiolabeled at the 5′ end of the pyrimidine strand Y*, is incubated at 37°C in 50 mM HEPES pH = 7.2, 100 mM NaCl, 10 mM MgCl2 in the presence of decreasing concentrations of: TFOs (1, 0.5, 0.3, 0.2 and 0.1 µM), TFO–TPT (2, 1, 0.8, 0.5 and 0.1 µM) and TFO–CPT (5, 2, 1 and 0.5 µM). Aliquots were removed after 2 h of incubation at 37°C and analyzed on a 15% nondenaturating acrylamide gel. (e) A radiolabeled 324-bp DNA fragment containing the duplex target (lane 1) was incubated at 37°C and pH 7.2 (28) in the presence of topoisomerase (lane 2) and of the TFO at 5 µM (lane 3), CPT at 10 µM (lane 4), TFO–CPT at 5 µM (lane 5), TPT at 10 µM (lane 6) and TFO–TPT at 5 µM (lane 7). Adenine/guanine-specific Maxam–Gilbert chemical cleavage reactions were used as markers (G + A). The position of the cleavage site of the conjugates is indicated by an arrow. The region corresponding to the triplex site is indicated (TFO), as the orientation.
Here, we make use of the ability to conjugate molecules of the camptothecin class to a TFO to directly probe the dynamical interactions between these antitumor drugs and TopIB at a single DNA sequence using single-molecule techniques. This use of TFO–CPT conjugates allows us to study the uncoiling process under well-defined conditions, i.e. at a predefined site on the DNA, with the expectation that the underlying physics on the slow uncoiling could be most effectively studied. Specifically, we measure the instantaneous uncoiling rate in the presence of drugs and the lifetime of the drug-stabilized covalent complex. By comparing these quantities to experiments performed with free TPT in solution, i.e. to sequence-independent uncoiling, we probe the effect of sequence on drug-mediated and TopIB-catalyzed supercoil removal. A previous single-molecule study (19) showed that the distribution in uncoiling velocities and lifetimes is rather broad, which led to the speculation that (part of) the variation in these quantities was caused by differing local interactions at the numerous sites at which uncoiling took place. We thus envisaged that confining the uncoiling to a unique site on the DNA might narrow the distribution of uncoiling velocities, which would enable the accurate study of the dependence of uncoiling velocity on parameters such as force and torque. In this way, our experiments allow us to determine whether sequence plays a dominant role in the dynamics of DNA uncoiling, which may help streamline the efforts currently underway to chemically improve the drugs.
MATERIALS AND METHODS
Synthesis of camptothecin and topotecan analogs
Mass determination was accomplished by electrospray ionization on a Q-STAR pulsar I (Appleura) and HPLC purifications were performed upon Agilent 1100 using a Xterra reverse phase C18 column (4.6 × 50 mm, 2.5 µm). 1H NMR were recorded in Me2SO-d6 on a Varian spectrometer (300 mHz). All the drugs were dissolved in Me2SO. 10-hydroxycamptothecin was purchased from Sigma and topotecan from Molekula.
Synthesis of 10-(6-aminohexyloxy)-topotecan NTPT (2):10-(6-Boc-aminohexyloxy)-topotecan
To a solution of 5 mg (11 µmol) topotecan hydrochloride in dry DMF (50 µl) was added diisopropylethylamine (4 µl; 44 µmol) and 6-(Boc-amino)hexyl bromide (10 µl; 44 µmol). The mixture was stirred at 40°C for 48 h, and then analyzed and purified by reverse phase HPLC using linear acetonitrile gradient (0–90% CH3CN with 0.1% of TFA) to afford a yellow solid (2.4 mg; 3.5 µmol; yield: 39%). ES-MS: found: (M + H) 620.33, calculated: 620.32 (C34H44N4O7). 1H NMR (Me2SO-d6): δ(p.p.m.) = 0.87(t, 3H, J = 7.2 Hz), 1.3–1.5 (m, 15 H), 1.8–1.9 (m, 4 H), 2.8–3 (m, 8 H), 4.31 (t, 2 H, J = 6.4 Hz), 4.7–4.8 (m, 2 H), 5.30 (s, 2 H), 5.43 (s, 2 H), 6.47 (br, 0.8 H), 7.31 (s, 1 H), 7.89–7.92 (d, 1 H, J = 9.5 Hz), 8.31–8.35 (d, 1 H, J = 9.5 Hz), 8.96 (s, 1 H).
10-(6-aminohexyloxy)-topotecan
The Boc-protected topotecan (2.8 mg; 4.6 µmol) was dissolved in CHCl3/TFA 1:1 and stirred at room temperature for 20 min (0.5 ml). The solvent was removed and the resulting product was purified by reverse phase HPLC using linear acetonitrile gradient (0–90% CH3CN with 0.1% of TFA) to afford a yellow solid (8.7 mg; 1.73 µmol; yield: 88%). ES-MS: found: (M + H) 520.27; calculated: 520.27 (C29H36N4O5). 1H NMR (Me2SO-d6): δ(p.p.m.) = 0.88(t, 3 H, J = 7.4 Hz), 1.36–1.60 (m, 6 H), 1.8–1.9 (m, 4 H), 2.7–2.9 (m, 8 H), 4.30 (t, 2 H, J = 6.4 Hz), 4.7–4.8 (m, 2 H) 5.30 (s, 2 H), 5.42 (s, 2 H), 6.50 (br, 1 H), 7.32 (s, 1 H), 7.68 (br, 2 H), 7.89–7.92 (d, 1 H, J = 9.5 Hz), 8.33–8.36 (d, 1 H, J = 9.5 Hz), 8.96 (s, 1 H). See Figure 1b for the detailed structure of this molecule.
Synthesis of 10-(6-aminohexyloxy)-camptothecin NCPT (4):10-(6-Boc-aminohexyloxy)-camptothecin
To a solution of 10 mg (27 µmol) 10-hydroxycamptothecin in dry acetone (10 ml) was added anhydrous potassium carbonate (4.5 mg; 32 µmol) and 6-(Boc-amino)hexyl bromide (11.3 mg; 30 µmol). The mixture was stirred at reflux for 18 h. The reaction mixture was filtered and the solvent removed. The resulting solid was analyzed and purified by reverse phase HPLC using linear acetonitrile gradient (0–90% CH3CN with 0.1% of TFA). This afforded a yellow solid (9.5 mg; 1.69 µmol; yield: 63%). ES-MS: found: (M + H) 563.62 Calculated: 563.64 (C31H37N3O7). 1H NMR (Me2SO-d6): δ(p.p.m.) = 0.87(t, 3 H, J = 7.3 Hz), 1.3–1.6 (m, 15 H), 1.79–2.07 (m, 4 H), 2.72–2.83 (m, 2 H), 4.13 (t, 2 H, J = 6.6 Hz), 5.27 (s, 2 H), 5.41 (s, 2 H), 6.47 (s, 1 H), 6.75 (m, 1 H), 7.28 (s, 1 H), 7.48–7.54 (m, 1 H), 8.06 (d, 1 H, J = 9.7 Hz), 8.52 (s, 1 H).
10-(6-aminohexyloxy)-camptothecin
The Boc-protected camptothecin (8 mg; 1.73 µmol) was dissolved in CHCl3/TFA 1:1 (1 ml) and stirred at room temperature for 2 h. The solvent was removed and the resulting product was purified by reverse phase HPLC using linear acetonitrile gradient (0–90% CH3CN with 0.1% of TFA) to afford a yellow/orange solid (8.7 mg; 1.73 µmol; yield: 90%). ES-MS: found: (M + H) 463.52; calculated: 463.53 (C26H29N3O5). 1H NMR (Me2SO-d6): δ(p.p.m.) = 0.88(t, 3 H, J = 7.3 Hz), 1.4–1.65 (m, 6 H), 1.79–2.26 (m, 4 H), 2.71–2.83 (m, 2 H), 4.15 (t, 2 H, J = 6.7 Hz), 5.27 (s, 2 H), 5.43 (s, 2 H), 6.40 (s, 1 H), 7.28 (s, 1 H), 7.48–7.54 (m, 1 H), 7.61 (m, 2 H), 8.05 (d, 1H, J = 9.7 Hz), 8.51 (s, 1 H). See Figure 1b for the detailed structure of this molecule.
General procedure for conjugation to the TFOs
Oligonucleotide TFO (sequence reported in Figure 1c) was purchased from Eurogentec and purified using quick spin columns and sephadex G-25 fine (Boehringer, Mannheim). Concentrations were determined spectrophotometrically at 25°C using molar extinction coefficients at 260 nm calculated from a nearest-neighbor model. The target DNA sequence is underlined in Figure 1c.
After precipitation as hexadecyltrimethylammonium salt 300 µg (58 µmol) of 3′-phosphorylated oligonucleotide (TFO) were dissolved in 45 μl of dry Me2SO. Solutions of 4-dimethylaminopyridine [5 mg (40 µmol) in 30 μl of Me2SO], dipyridyl disulfide (6.6 mg in 30 μl of Me2SO, 30 μmol) and triphenylphospine (8 mg in 30 μl of Me2SO, 30 μmol) were added. After 15 min of incubation at room temperature, the activated oligonucleotide was precipitated with 2% LiClO4 in acetone, rinsed with acetone and dissolved in a solution of 30 µl of Me2SO/water 8:2 of the amino-derivative (0.5 µmol). After 2 h of incubation, the oligonucleotide conjugate was precipitated with 2% LiClO4 in acetone, rinsed with acetone and purified by reverse phase HPLC using linear acetonitrile gradient [0–80% CH3CN in 0.2 M (NH4)OAc]. TFO–TPT (yield 74%): ES-MS: found: (M + H) 5646.3 calculated: 5646.2. HPLC retention time: 12.9 min. TFO–CPT (yield 69%): ES-MS: found: (M + H) 5589.0 calculated: 5590.2. HPLC retention time: 12.4 min.
TFO–TPT and TFO–CPT binding to dsDNA
We show that the TFO–TPT and TFO–CPT conjugates bind to dsDNA and form a triple helix at pH 7.2 and 37°C, 100 mM NaCl and 10 mM MgCl2. The binding is observed by a decrease in the mobility on a nondenaturing polyacrylamide gel with increasing conjugate concentrations (Figure 1d). The radiolabeled oligopyrimidine strand of the duplex and the radiolabeled duplex were used as markers. The experimental procedure is described in ref. 28. Subsequent sequence-specific DNA cleavage by the conjugates is described in the Results section and Figure 1e.
Single-molecule experimental configuration and buffer conditions
Our experimental design, the magnetic tweezers, consists of a pair of magnets mounted above a flow cell of two glass slides with parafilm spacing (30,31). A linear continuous single dsDNA molecule with a length of ∼19.2 kb is tethered at its biochemically modified ends between a paramagnetic bead and the bottom glass slide using standard streptavidin-biotin (bead) and digoxygenin-anti-digoxigenin (surface) interactions. The central portion of DNA contains part of a Supercos1 plasmid (Stratagene), while the ends are 1238-bp stretches of pbluescriptIISK+ plasmid (Stratagene) obtained using a PCR reaction with biotinylated or digoxigenin-labeled nucleotides. Twisting the magnets about their center axis twists the DNA molecule, while translating the magnets in the vertical direction modulates the stretching force F on the DNA.
To perform single-molecule experiments on DNA molecules containing a TFO, the triple helix was first preformed inside the flow cell by flushing in a buffer containing 50 mM Tris–HCl (pH 7.2), 10 mM MgCl2, 60 mM KCl, 0.5 mM DTT, 200 μg/ml BSA, 0.1 mM EDTA and 10 μM TFO. The TFO was incubated for 90 min at room temperature. Experiments were performed in a buffer containing 50 mM Tris–HCl (pH 7.2), 10 mM MgCl2, 60 mM KCl, 0.5 mM DTT, 200 μg/ml BSA, 0.1% Tween. Under these conditions, the only TPT molecules present in the flow cell are those that are conjugated to the TFO. As we observe TPT-mediated slow supercoil removal in these buffer conditions, we can be sure that the triple helix remains formed during the course of the experiments.
RESULTS
Synthesis and binding of conjugates
The synthesis of the TFO conjugate of topotecan (TFO–TPT; Figure 1a) required the preparation of 10-(6-aminohexyloxy)-topotecan (NTPT). In the remainder of the text, NTPT and NCPT are referred to as TPT and CPT, respectively. The synthesis was carried out as outlined in Figure 1b (see also Materials and Methods section). The substitution of the phenolic OH was a critical step and mild conditions (40°C for 48 h) were used in order to avoid degradation of topotecan. Deprotection in TFA yielded the TPT in 35% yield. For comparison the same linker arm (6-aminohexyloxy) was attached to 10-hydrocamptothecin following standard procedures (CPT: 84% yield). The two amino-derivatives (2 and 4) were attached to the 3′ phosphate group of the TFO as described to obtain conjugates TFO–TPT and TFO–CPT.
Triplex formation by the conjugates was attested by PAGE (Figure 1d, Materials and Methods section). As expected, the presence of the drug destabilizes triplex formation, however the triple helix is still strongly formed at 37°C in the presence of 10 mM MgCl2 (C50 = 0.28, 0.75, 0.83 µM for TFO, TFO–TPT and TFO–CPT, respectively). In vitro cleavage experiments confirm that the conjugate induced TopIB-mediated DNA cleavage at a single site only (Figure 1e), located at the 3′ end of the triplex site where the drug is positioned by triplex formation (Figure 1c). We thus conclude that TFO–TPT binds to dsDNA and induces TopIB-mediated DNA cleavage at a predefined position. The cleavage obtained in the presence of TFO–CPT is seven times stronger than the one obtained in the presence of TFO–TPT. This difference is presumably due to a better fit by the TFO ligand of the CPT derivative in the ternary complex than the TPT one.
The TFO does not influence the mechanical properties of the DNA
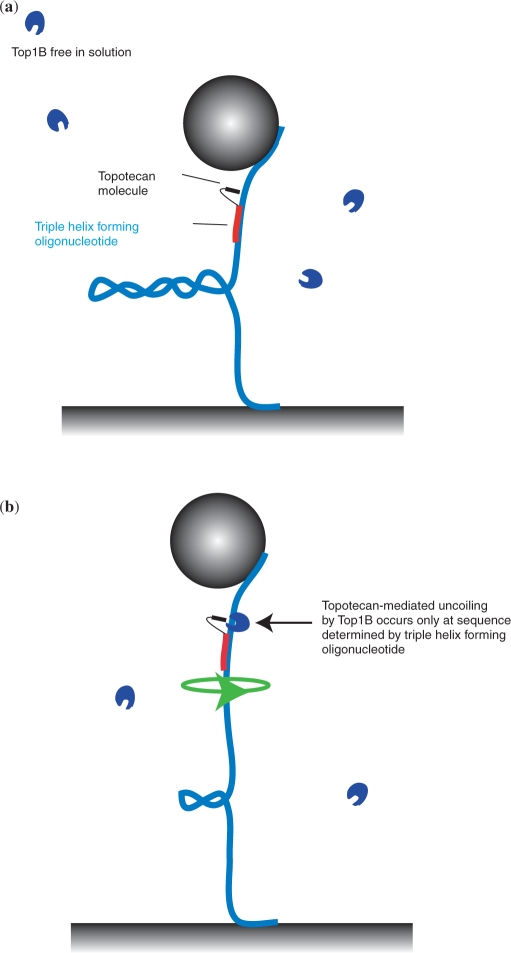
Our experimental configuration, the magnetic tweezers, is described elsewhere (30,31) and in brief consists of monitoring the length of a tethered DNA molecule in a flow cell above which a pair of magnets is mounted (see Materials and Methods section). Upon twisting the molecule via magnet rotation, plectonemic supercoils form (Figure 2a), causing the height of the bead above the surface to decrease. When TopIB (blue C-shaped structures) is added in combination with the TFO–TPT (TFO shown in red, TPT in black), the plectonemes will be removed (Figure 2b). This can occur either in a TPT-dependent manner, in which case the DNA sequence at which the uncoiling occurs is known, or in a TPT-independent manner, which occurs at an unknown sequence.
Figure 2.
Experimental strategy to detect sequence-specific uncoiling by TopIB at the single-molecule level. (a) A supercoiled double-stranded and continuous DNA molecule (blue) is tethered between a magnetic bead (gray sphere) and a glass surface. The TFO (red), connected to the TPT (black) binds in a sequence-specific manner to the DNA molecule. TopIB enzyme molecules (blue C-shaped structures) diffuse freely in buffer solution. (b) TopIB can only remove DNA supercoils in a drug-dependent manner at a site on the DNA that is dictated by the sequence-specific binding of the TFO–TPT.
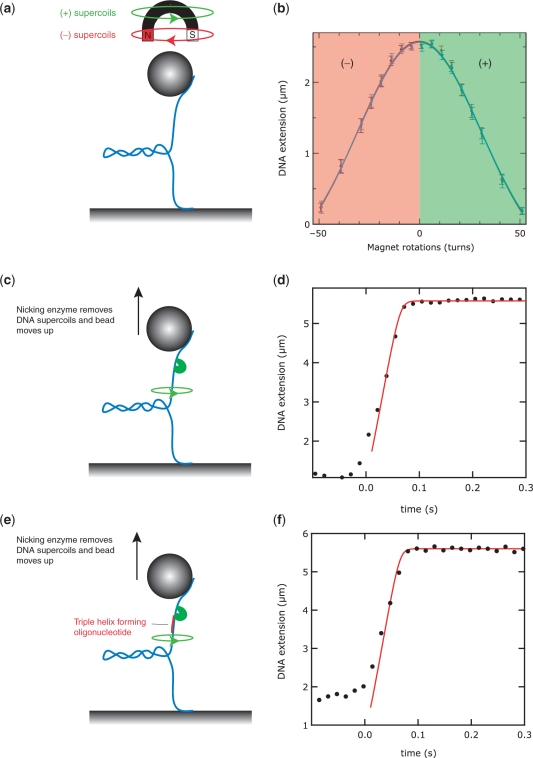
We performed several control experiments to ensure that the binding of the TFO–TPT did not influence the mechanical properties of the DNA molecule. As this study examines the enzymatic relaxation of plectonemic supercoils in the presence of a TFO, we first probed the influence of TFO on the mechanical coiling and the formation of plectonemic supercoils. To this end, we applied twist to the DNA molecule by rotating the magnets, either in the presence or absence of a bound triple helix. We then tested whether plectonemic supercoils formed at the same degree of twist and whether the supercoils that formed were of similar dimensions. Rotating the magnets about their axis in the direction of the natural helicity of the DNA induces overwinding of the DNA molecule that will lead to buckling of the DNA and the formation of positive plectonemic supercoils [Figure 3a and (32)]. Rotating in the opposite direction leads to underwinding and the formation of negative supercoils in the DNA. Figure 3b plots the DNA extension as a function of the number of magnet rotations. At low stretching force (F smaller than ∼0.5 pN, depending on buffer conditions), the DNA's response to twist is symmetrical for positive and negative supercoils and is identical for DNA without TFO bound (red data points) and for DNA with TFO bound (blue data points). Error bars denote the standard deviation of the bead fluctuations.
Figure 3.
The presence of the TFO does not influence quasi-static mechanical properties of DNA. (a) Using the magnetic tweezers, positive (+) or negative (–) supercoils can be readily introduced in the DNA molecule by either twisting the magnets along with (green arrow) or against (red arrow) the natural helicity of the DNA. (b) Under the appropriate conditions of stretching force and buffer, the response of the DNA molecule to twisting is symmetrical: both (+) and (–) supercoils cause the DNA extension to decrease in an equal manner, as plectonemic supercoils form. (c) A positively supercoiled molecule will relax, when cleaved by a nicking enzyme. Consequently, the DNA extension will increase in time. (d) The dynamics of the uncoiling is in thermal equilibrium, and is well-predicted by a model that is described in ref. 33. (e) Cleavage of a supercoiled DNA molecule, in the presence of a bound TFO construct. The sequence of the TFO is chosen such that cleavage occurs at a distance of 5 bp from the 3′-end of the TFO. (f) The dynamics of supercoil removal after cleavage with a nicking enzyme in the presence of TFO is well described by the quasi-static model, similar to (d).
We also asked whether a bound TFO would influence the swivelling of a supercoiled DNA molecule after cleavage by an enzyme. Torque stored in the supercoiled DNA will dissipate through the nick upon cleavage by topoisomerase or a nicking enzyme. As a result, the DNA will start to spin (green arrow in Figure 3c), plectonemic supercoils are removed and the bead will move upwards. The dynamics of DNA extension following cleavage by a nicking enzyme has previously been quantitatively studied and modeled (33), allowing us to rapidly assess the influence of the TFO on this process. The nicking enzyme and the TopIB used in subsequent experiments cut the DNA at comparable distances from the 3′-end of the triplex (5 bp versus 4 bp, respectively). The DNA extension trajectory in time after cleavage by a nicking enzyme (at time t = 0 s) is plotted in Figure 3d (black dots are samplings of the bead height at 60 Hz). The red solid line in Figure 3d is the prediction of a quasi-static model for the stretching dynamics of DNA that takes into account the constant stretching force pulling at the DNA, the Stokes drag on the bead as it moves through the water, and the entropic cost for extending a torsionally relaxed worm-like chain such as DNA (33). The DNA extension saturates at the equilibrium extension, which is a function of F, of the crystallographic length of the DNA, and of the persistence length of DNA, which we have independently measured to be 53 ± 2 nm, in agreement with literature (30). In the presence of TFO and a nicking enzyme, the DNA extension trajectory in time is equally well described by the model (Figure 3e and f). We thus conclude that the presence of the TFO does not influence the quasi-static mechanical properties of the DNA, or the uncoiling dynamics upon enzymatic cleavage at a locus close to the TFO.
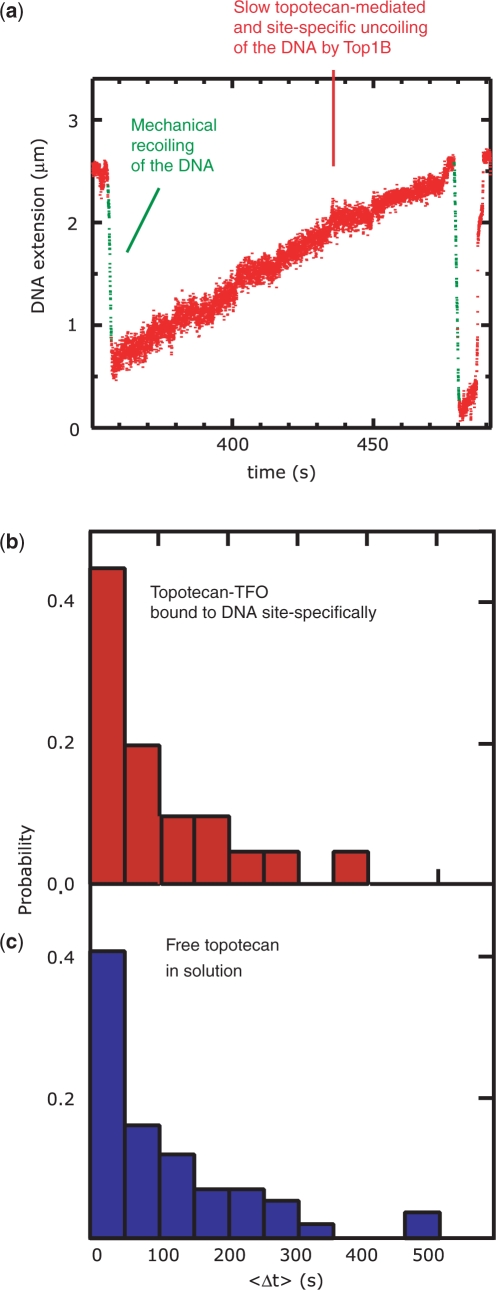
Drug-mediated slow uncoiling and lifetime of the covalent complex
Previous experiments have shown that in the presence of TopIB, the plectonemes in a mechanically supercoiled DNA molecule are enzymatically removed at a low rate if the uncoiling is mediated by TPT (19). In summary, for the removal of positive supercoils this rate was typically found to be roughly 20 times lower in comparison to the rate of non-TPT-mediated supercoil removal and is only observed in the presence of a catalytically active human TopIB. Furthermore, the rate of DNA religation is decreased ∼400-fold (19). Thus, slow supercoil removal constitutes a signature for TPT-mediated uncoiling and can be used to measure the lifetime of the covalent complex. Here we extend these measurements by investigating the effect of DNA sequence using the TFO–TPT construct. With a bound TFO–TPT and in the presence of TopIB, slow supercoil removal can be observed (red data points in Figure 4a), similar to experiments with free TPT (19). We note that with free TPT in solution as well as with the TFO–TPT construct, most uncoiling events are not TPT-mediated. However, with the TFO–TPT construct, the relative frequency of TPT-mediated uncoiling events is even lower (less than ∼10%), in agreement with the restriction that this experimental design imposes on the number of available DNA loci for TPT-mediated uncoiling. The time duration of slow supercoil removal, Δt, was measured and the outcomes are displayed in a histogram, both for the TFO–TPT construct (sequence-specific uncoiling, Figure 4b, <Δt> = 115 s) and free TPT in solution (nonsequence-specific uncoiling, Figure 4c, <Δt>=124 s). It is apparent from Figure 4b and c that the distributions do not differ significantly. Earlier biochemical studies have shown that camptothecin stabilizes the covalent TopIB–DNA intermediate at a number of different sites on the DNA to different degree (12,14). We thus expected that in the context of single-molecule measurements, the broadness of the Δt-distribution for free TPT in solution (Figure. 4c) might be caused by the heterogeneity in the degree to which different sites on the DNA are stabilized. However, we find that the broadness of the Δt-distribution for a unique DNA sequence, i.e. in the TFO–TPT measurement, is roughly as broad as the Δt-distribution for multiple sites, i.e. in the free TPT measurement. We therefore conclude that the variation in Δt is not caused by variation in DNA sequence at which uncoiling occurs. There is, however, one relevant caveat in the measurement of Δt, which is that this number may constitute a lower bound (19). This is because the transition from slow uncoiling to fast uncoiling, which is used as the criterion for the time up to which slow supercoiling persists, can be explained in two different ways. First, this transition can be caused by TPT exiting the covalent complex, in which case the measurement of Δt is a correct measure of the lifetime of the covalent complex [religation is fast compared to Δt, (19)]. The second explanation is that during slow TPT-mediated supercoil removal by a first TopIB, a second, independent TopIB cleaves the DNA at another site on the DNA in a TPT-independent manner, giving rise to fast supercoil removal. In this case, Δt is a measure for the time between independent cleavage events by different TopIBs, and represents a lower bound for the lifetime of the covalent complex. In principle, varying the TopIB concentration in the flow cell ought to exclude the second explanation. We have varied the concentration by a factor of 2, which is the largest concentration range experimentally feasible (6) and have seen no effect on the Δt distribution (19). However, we cannot be fully confident that the TopIB concentration in the bulk solution changes as expected given the large surface-to-volume ratio of the flow cell and the tendency of TopIB to bind to the negatively charged polyglutamic acid used to passivate the flow cell surfaces. We can nonetheless conclude that restricting the formation of a TPT-stabilized covalent complex to a specific sequence does not dramatically alter its lifetime in comparison to the situation in which the complex is allowed to form at an ensemble of sites on the DNA.
Figure 4.
Lifetime of the covalent TopIB–DNA complex. (a) The DNA molecule is mechanically coiled by twisting the magnets, leading to a decrease in DNA extension (green lines). In this trace, positive supercoils are immediately removed by TopIB in a TPT-dependent fashion, with TPT bound to the TFO. Supercoil removal manifests itself as a slow increase in DNA extension (red dots). The total time duration of slow continuous supercoil removal, <Δt>, is measured for a number of uncoiling events for both sequence-specific uncoiling using the TFO–TPT construct (b, red histogram, n = 20) and for nonsequence-specific uncoiling using free TopIB in solution (c, blue histogram, n = 122). The distributions have similar widths.
Measurement of the uncoiling velocities of positive and negative supercoils
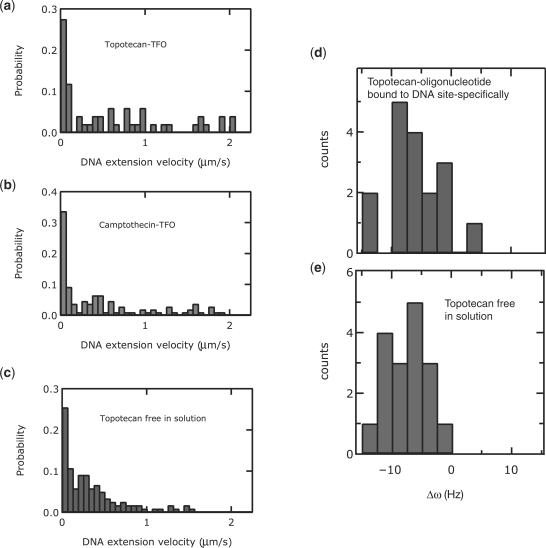
To quantify the rate of drug-mediated supercoil removal by TopIB at a single DNA sequence, we measured the distribution of uncoiling velocities during supercoil removal in the presence of the TFO–TPT (Figure 5a, <v> = 0.5 ± 0.1 μm/s) and TFO–CPT (Figure 5b, <v> = 0.4 ± 0.1 μm/s) constructs. These were compared to the measurements in the presence of free TPT (Figure 5c, <v> = 0.3 ± 0.1 μm/s). Here, a velocity of 2.1 μm/s was used as a cutoff to define TPT-mediated events, and all errors indicated are standard deviations. We also measured the asymmetry in uncoiling rates Δω = ω+ – ω–(19) in the presence of a bound TFO–TPT (Figure 5d, < Δω> = –8 ± 6 Hz) and for free TPT (Figure 5e, <Δω> = –8 ± 5 Hz). What is apparent from these histograms is that the distributions are roughly equally broad in the case of sequence-specific uncoiling in comparison to sequence-independent uncoiling. One might expect that the local microscopic environment, i.e. the molecular interactions between the drugs, the TopIB and the DNA would be of such importance that the effect of sequence would manifest itself in our sensitive single-molecule measurements. In such a case, we would specifically have predicted that confining the drug-mediated uncoiling to a unique site would lead to a narrowing of the distribution of uncoiling velocities. This prediction was not borne out in our measurements and we conclude that sequence does not play a dominating role in the velocity of drug-mediated DNA uncoiling. Thus, while our single-molecule experiments can sensitively detect the altered dynamics of supercoil removal as a result of subtle effects such as a change in the helicity of uncoiling, they reveal that the role of DNA sequence in the drug-mediated TopIB dynamics is in comparison relatively minor.
Figure 5.
Instantaneous uncoiling velocity in the presence of TPT. Uncoiling velocities in the presence of the camptothecins. (a), (b) and (c) show histograms of instantaneous uncoiling velocities of TFO–TPT ((a), n = 51, red histogram), TFO-CPT ((b), n = 110, green histogram) and free TPT in solution (C, n = 122, blue histogram). Histograms shown in (a), (b) and (c) are all similar in their means and width. (d) histogram of the asymmetry in uncoiling velocities of the removal of negative and positive supercoils, in the presence of TFO–TPT (n = 17, red histogram). Δω is a measure for the differential in uncoiling rates: Δω = ω+ – ω–, where ω+ is the uncoiling rate of positive supercoils and ω– is the uncoiling rate of negative supercoils. (e) Histogram of the asymmetry in uncoiling velocities of the removal of negative and positive supercoils in the presence of free TPT in solution (n = 17, blue histogram). Histograms shown in (d) and (e) are similar in their means and width.
DISCUSSION
We have experimentally demonstrated topotecan- and camptothecin-mediated slow supercoil removal by human topoisomerase IB restricted to a specific site on the DNA. Our main conclusion is that the dynamics of slow uncoiling observed at a single site is indistinguishable from the slow uncoiling we observe on DNA molecules where multiple sites are available for drug binding. The experimental observables that showed no difference were the instantaneous rate of positive and negative supercoil removal, as well as the lifetime of the drug-stabilized covalent TopIB–DNA complex. In the context of these measurements, we raise a number of points for discussion.
First, the lifetime of the covalent complex in the presence of drug is about 2 min, both for sequence-specific and nonsequence-specific uncoiling (19). The observation that these two quantities are identical for all practical purposes indicates that the measurement of the lifetime for free TPT could not have been overestimated due to a potential limitation to distinguish two subsequent and independent TopIB enzymes uncoiling in the presence of drug at two different sites on the DNA. In such a scenario, a recorded event of long uncoiling could in reality consist of two events that are somewhat shorter in duration, but overlap in time.
Second, it may still be possible that ‘the same’ TPT molecule enters and exits the TopIB–DNA complex on a time scale that is faster than our time resolution. However, we note that this is not a particularly relevant caveat from a biological point of view, as such fast dynamics still lead to slow DNA uncoiling, which is what the cell and its DNA processing apparatus, and in particular the replication fork, has to confront.
Third, our data suggest that DNA sequence does not influence the uncoiling rate and the lifetime of the drug-stabilized covalent complex. It might have been expected that the exact chemical structure at the swivel point of the TopIB would influence the swivel rate. Similarly, it might have been expected that the interactions between the TopIB, the DNA and TPT would have an effect on the binding strength of TPT inside the enzyme cavity. Both of these speculations are not borne out by our measurements. Rather, the broad distributions that our single-molecule measurements reveal may have to be attributed to stochastic variations in the binding of the TPT into the pocket formed by the TopIB and the nicked DNA, stochastic variations in the manner that TopIB encircles the DNA, or, in the case of the lifetimes of the TPT complex, variable breathing modes of either the TopIB or the DNA molecule itself.
The outcome of our dynamical experiments combined with the relative sequence independence of DNA cleavage (25) implies that sequence may not play an important role in considerations of drug efficacy. Particularly if drug targeting proceeds via TFOs, the observed sequence-independent dynamics suggest that such conjugates can be used as tools to direct the action of camptothecin to a specific site (e.g. on a specific gene in the human genome) without altering the poisoning features of the drug. Our findings have an impact on the process of designing improved camptothecins. Chemical modification of the TopIB (34) or camptothecins (35) appear to affect the TopIB reaction kinetics to a large degree, but our data suggest that the DNA does not need to be taken into account in the design process. This may pose a potential simplification in the strategy to target DNA sequences specific to certain cancers, since the overall strategy need not be adapted on a case-by-base basis.
ACKNOWLEDGEMENTS
The authors thank Cees Dekker, Mary-Ann Bjornsti, Ulrich Schwarz, and Sidney Hecht for useful discussions, and Lionel Dubost and Alain Blond of mass and NMR facilities of the MNHN for technical help. The authors also thank Brigitta Witte, Susanne Hage and Ya-Hui Chien for help in synthesizing and characterizing DNA constructs for use in the tweezer experiments. This work was supported by grants from Ligue Nationale Contre le Cancer and NWO. Funding to pay the Open Access publication charges for this article was provided by NWO.
Conflict of interest statement. None declared.
REFERENCES
- 1.Champoux JJ. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 2.Wang JC. DNA topoisomerases. Annu. Rev. Biochem. 1996;65:635–692. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- 3.Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell. Biol. 2002;3:430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 4.Corbett KD, Berger JM. Structure, molecular mechanisms, and evolutionary relationships in DNA topoisomerases. Annu. Rev. Biophys. Biomol. Struct. 2004;33:95–118. doi: 10.1146/annurev.biophys.33.110502.140357. [DOI] [PubMed] [Google Scholar]
- 5.Kim RA, Wang JC. Function of DNA topoisomerases as replication swivels in Saccharomyces cerevisiae. J. Mol. Biol. 1989;208:257–267. doi: 10.1016/0022-2836(89)90387-2. [DOI] [PubMed] [Google Scholar]
- 6.Moreno-Herrero F, Holtzer L, Koster DA, Shuman S, Dekker C, Dekker NH. Atomic force microscopy shows that vaccinia topoisomerase IB generates filaments on DNA in a cooperative fashion. Nucleic Acids Res. 2005;33:5945–5953. doi: 10.1093/nar/gki906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sekiguchi J, Shuman S. Vaccinia topoisomerase binds circumferentially to DNA. J. Biol. Chem. 1994;269:31731–31734. [PubMed] [Google Scholar]
- 8.Koster DA, Croquette V, Dekker C, Shuman S, Dekker NH. Friction and torque govern the relaxation of DNA supercoils by eukaryotic topoisomerase IB. Nature. 2005;434:671–674. doi: 10.1038/nature03395. [DOI] [PubMed] [Google Scholar]
- 9.Stivers JT, Harris TK, Mildvan AS. Vaccinia DNA topoisomerase I: evidence supporting a free rotation mechanism for DNA supercoil relaxation. Biochemistry. 1997;36:5212–5222. doi: 10.1021/bi962880t. [DOI] [PubMed] [Google Scholar]
- 10.Stewart L, Redinbo MR, Qiu X, Hol WG, Champoux JJ. A model for the mechanism of human topoisomerase I. Science. 1998;279:1534–1541. doi: 10.1126/science.279.5356.1534. [DOI] [PubMed] [Google Scholar]
- 11.Bjornsti MA. Cancer therapeutics in yeast. Cancer Cell. 2002;2:267–273. doi: 10.1016/s1535-6108(02)00160-5. [DOI] [PubMed] [Google Scholar]
- 12.Li TK, Liu LF. Tumor cell death induced by topoisomerase-targeting drugs. Annu. Rev. Pharmacol. Toxicol. 2001;41:53–77. doi: 10.1146/annurev.pharmtox.41.1.53. [DOI] [PubMed] [Google Scholar]
- 13.Minsky BD. Combined-modality therapy of rectal cancer with irinotecan-based regimens. Oncology. 2004;18:49–55. [PubMed] [Google Scholar]
- 14.Pommier Y. Topoisomerase I inhibitors: camptothecins and beyond. Nat. Rev. Cancer. 2006;6:789–802. doi: 10.1038/nrc1977. [DOI] [PubMed] [Google Scholar]
- 15.Rodriguez-Galindo C, Radomski K, Stewart CF, Furman W, Santana VM, Houghton PJ. Clinical use of topoisomerase I inhibitors in anticancer treatment. Med. Pediatr. Oncol. 2000;35:385–402. doi: 10.1002/1096-911x(20001001)35:4<385::aid-mpo1>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 16.Stewart DJ. Topotecan in the first-line treatment of small cell lung cancer. Oncologist. 2004;9(Suppl. 6):33–42. doi: 10.1634/theoncologist.9-90006-33. [DOI] [PubMed] [Google Scholar]
- 17.Chrencik JE, Staker BL, Burgin AB, Pourquier P, Pommier Y, Stewart L, Redinbo MR. Mechanisms of camptothecin resistance by human topoisomerase I mutations. J. Mol. Biol. 2004;339:773–784. doi: 10.1016/j.jmb.2004.03.077. [DOI] [PubMed] [Google Scholar]
- 18.Hsiang YH, Hertzberg R, Hecht S, Liu LF. Camptothecin induces protein-linked DNA breaks via mammalian DNA topoisomerase I. J. Biol. Chem. 1985;260:14873–14878. [PubMed] [Google Scholar]
- 19.Koster DA, Palle K, Bot ES, Bjornsti MA, Dekker NH. Antitumour drugs impede DNA uncoiling by topoisomerase I. Nature. 2007;448:213–217. doi: 10.1038/nature05938. [DOI] [PubMed] [Google Scholar]
- 20.Porter SE, Champoux JJ. The basis for camptothecin enhancement of DNA breakage by eukaryotic topoisomerase I. Nucleic Acids Res. 1989;17:8521–8532. doi: 10.1093/nar/17.21.8521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Staker BL, Hjerrild K, Feese MD, Behnke CA, Burgin AB, Jr, Stewart L. The mechanism of topoisomerase I poisoning by a camptothecin analog. Proc. Natl Acad. Sci. USA. 2002;99:15387–15392. doi: 10.1073/pnas.242259599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holm C, Covey JM, Kerrigan D, Pommier Y. Differential requirement of DNA replication for the cytotoxicity of DNA topoisomerase I and II inhibitors in Chinese hamster DC3F cells. Cancer Res. 1989;49:6365–6368. [PubMed] [Google Scholar]
- 23.Hsiang YH, Lihou MG, Liu LF. Arrest of replication forks by drug-stabilized topoisomerase I-DNA cleavable complexes as a mechanism of cell killing by camptothecin. Cancer Res. 1989;49:5077–5082. [PubMed] [Google Scholar]
- 24.Jaxel C, Capranico G, Kerrigan D, Kohn KW, Pommier Y. Effect of local DNA sequence on topoisomerase I cleavage in the presence or absence of camptothecin. J. Biol. Chem. 1991;266:20418–20423. [PubMed] [Google Scholar]
- 25.Matteucci M, Lin K-Y, Huang T, Wagner R, Sternbach DD, Mehrotra M, Besterman JM. Sequence-specific targeting of duplex DNA using a camptothecin-triple helix forming oligonucleotide conjugate and topoisomerase I. J. Am. Chem. Soc. 1997;119:6939–6940. [Google Scholar]
- 26.Hurley LH. DNA and its associated processes as targets for cancer therapy. Nat. Rev. Cancer. 2002;2:188–200. doi: 10.1038/nrc749. [DOI] [PubMed] [Google Scholar]
- 27.Arimondo PB, Bailly C, Boutorine AS, Ryabinin VA, Syniakov AN, Sun JS, Garestier T, Helene C. Directing topoisomerase I mediated DNA cleavage to specific sites by camptothecin tethered to minor- and major-groove ligands. Angew. Chem. Int. Ed. Engl. 2001;40:3045–3048. doi: 10.1002/1521-3773(20010817)40:16<3045::AID-ANIE3045>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 28.Arimondo PB, Boutorine A, Baldeyrou B, Bailly C, Kuwahara M, Hecht SM, Sun JS, Garestier T, Helene C. Design and optimization of camptothecin conjugates of triple helix-forming oligonucleotides for sequence-specific DNA cleavage by topoisomerase I. J. Biol. Chem. 2002;277:3132–3140. doi: 10.1074/jbc.M110181200. [DOI] [PubMed] [Google Scholar]
- 29.Arimondo PB, Thomas CJ, Oussedik K, Baldeyrou B, Mahieu C, Halby L, Guianvarc'h D, Lansiaux A, Hecht SM, Bailly C, et al. Exploring the cellular activity of camptothecin-triple-helix-forming oligonucleotide conjugates. Mol. Cell. Biol. 2006;26:324–333. doi: 10.1128/MCB.26.1.324-333.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bustamante C, Bryant Z, Smith SB. Ten years of tension: single-molecule DNA mechanics. Nature. 2003;421:423–427. doi: 10.1038/nature01405. [DOI] [PubMed] [Google Scholar]
- 31.Strick T, Allemand J, Croquette V, Bensimon D. Twisting and stretching single DNA molecules. Prog. Biophys. Mol. Biol. 2000;74:115–140. doi: 10.1016/s0079-6107(00)00018-3. [DOI] [PubMed] [Google Scholar]
- 32.Strick TR, Allemand JF, Bensimon D, Bensimon A, Croquette V. The elasticity of a single supercoiled DNA molecule. Science. 1996;271:1835–1837. doi: 10.1126/science.271.5257.1835. [DOI] [PubMed] [Google Scholar]
- 33.Crut A, Koster DA, Seidel R, Wiggins CH, Dekker NH. Fast dynamics of supercoiled DNA revealed by single-molecule experiments. Proc. Natl Acad. Sci. USA. 2007;104:11957–11962. doi: 10.1073/pnas.0700333104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gao R, Zhang Y, Dedkova L, Choudhury AK, Rahier NJ, Hecht SM. Effects of modification of the active site tyrosine of human DNA topoisomerase I. Biochemistry. 2006;45:8402–8410. doi: 10.1021/bi0605179. [DOI] [PubMed] [Google Scholar]
- 35.Hecht SM. Camptothecin: roles of the D and E rings in binding to the topoisomerase I-DNA covalent binary complex. Curr. Med. Chem. Anticancer Agents. 2005;5:353–362. doi: 10.2174/1568011054222373. [DOI] [PubMed] [Google Scholar]