Abstract
let-7 microRNA (miRNA) regulates heterochronic genes in developmental timing of the nematode Caenorhabditis elegans. Binding of miRNA to messenger RNA (mRNA) and structural features of the complex are crucial for gene silencing. We herein present the NMR solution structure of a model mimicking the interaction of let-7 miRNA with its complementary site (LCS 2) in the 3′ untranslated region (3′-UTR) of the lin-41 mRNA. A structural study was performed by NMR spectroscopy using NOE restraints, torsion angle restraints and residual dipolar couplings. The 33-nt RNA construct folds into a stem–loop structure that features two stem regions which are separated by an asymmetric internal loop. One of the stems comprises a GU wobble base pair, which does not alter its overall A-form RNA conformation. The asymmetric internal loop adopts a single, well-defined structure in which three uracils form a base triple, while two adenines form a base pair. The 3D structure of the construct gives insight into the structural aspects of interactions between let-7 miRNA and lin-41 mRNA.
INTRODUCTION
MicroRNAs (miRNAs) represent a large class of small RNAs that function as negative gene regulators in eukaryotes (1–3). They regulate diverse biological processes such as developmental timing, differentiation, cell proliferation and apoptosis. Bioinformatics data indicate that each miRNA can control hundreds of gene targets, underscoring the potential influence of miRNAs on many genetic pathways. The enzyme Dicer produces miRNAs from endogenous stem–loop RNA molecules giving rise to single-stranded molecules of about 21 to 23 nt in length (4). Mature functional miRNA molecules are partially complementary to mRNA molecules (5) and their function is to repress protein synthesis. An example of an evolutionary conserved miRNA is let-7 (6). The 21-nt let-7 miRNA, which was originally discovered as an essential regulator of developmental timing in the nematode Caenorhabditis elegans, was later shown to be highly conserved in several organisms including humans (7,8). On the other hand, lin-41 is the first C. elegans heterochronic gene that has mammalian homologs involved in developmental timing. Studies by Slack et al. (9) showed that lin-41 is temporarily expressed during mouse embryonic development in several tissues (9). While expression of lin-41 is downregulated during mouse development, expression of let-7 is upregulated. lin-41 and many other mRNAs possess complementary sites for let-7 miRNA in their 3′-UTR regions. Numerous miRNAs, termed ‘oncomirs’ are associated with cancer (10). In this respect, it has recently emerged that let-7 is a promising therapeutic agent to treat lung cancer caused by mutations in RAS lung genes (11). The formation of let-7:lin-41 complexes suppresses expression of lin-41 through the miRNA silencing mechanism which in turn regulates the transition from the last larval stage to adulthood in a nematode.
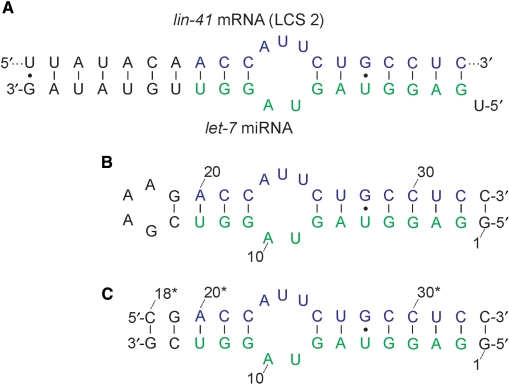
The purpose of the current study was to expand our knowledge on the process of miRNA-induced control of gene expression by focusing on the structural determinants that influence the stability of let-7 miRNA:lin-41 mRNA complex with the use of high-resolution NMR spectroscopy in aqueous solution. let-7 miRNA forms two distinct complexes with the 3′-UTR of the lin-41 mRNA (6). We have concentrated on one of the let-7 complementary sites (LCS 2) and designed a 33-nt model RNA stem–loop construct by linking the two strands (i.e. miRNA and mRNA) with the GAAA tetraloop (Figure 1). Our construct forms a stable structure consisting of two stem regions separated by the asymmetric internal loop. An additional CG base pair was included to close the GAAA tetraloop and increase the stability of the construct. 5′-U of the let-7 miRNA was removed, and an additional GC base pair was added at the end of the construct to facilitate in vitro transcription.
Figure 1.
(A) Schematic representation of the complex of let-7 miRNA with 3′-UTR of the lin-41 mRNA (LCS 2). The residues colored in green and blue match the sequences used in the monomolecular and dimeric constructs. (B) The 33-nt monomolecular RNA construct mimicking the complex. (C) The dimeric RNA construct. Numbering used in (B) has been preserved to facilitate the comparison between the two constructs. The residues originating from the lin-41 mRNA sequence are labeled by asterisk.
The 3D structure of the miRNA:mRNA construct was expected to give new insights into the structural aspects of interactions between a small noncoding RNA molecule (let-7) and respective mRNA (3′-UTR of the lin-41). In this way, a potential role of the internal loops and mismatch base pairs which are frequently found in miRNA:mRNA complexes involved in the regulation of gene expression would be evaluated. Elements of secondary structure other than canonical Watson–Crick base pairs are expected to potentially induce bends in the structure or lead to changes in groove dimensions which are important for recognition in the RISC complex (12). These local structural changes are expected to affect the thermodynamic stability of the miRNA:mRNA complex, water and cation localization, its predisposition for interaction with other (macro)molecules and molecular recognition properties. Similarly, it is not clear what is the limiting degree and nature of base pairing that is decisive as to choice of the gene suppression pathway entered by small noncoding RNAs. It has been shown that miRNAs can also work as small interfering RNAs (siRNAs) which are perfectly base paired with target mRNAs and negatively regulate gene expression by promoting degradation of mRNAs (13).
MATERIALS AND METHODS
Sample preparation
The 13C,15N uniformly labeled RNA construct was transcribed using standard enzymatic methods (14) with T7 RNA polymerase (Promega), 13C,15N-labeled rNTPs (Silantes) and a partially double-stranded DNA template (IDT-DNA) consisting of a T7 promoter and a coding template strand. The template strand was modified with C2′-methoxyls on the last two residues of the 5′ end (15). The transcription reaction was quenched using EDTA, the transcribed RNA was precipitated in ethanol and the precipitate was recovered after centrifugation by redissolving it in water. The target 33-nt RNA sequence was purified from abort transcripts using denaturating (7 M urea) PAGE electrophoresis (17%). The transcribed 33-nt RNA was excised from the gel and recovered by electroelution (Schleicher & Schuell). The electroeluted RNA solution was finally extensively dialyzed against an NMR buffer (starting from a high salt buffer and gradually reducing the ionic strength) using microcentrifugation tubes (Centricon, Millipore). The NMR sample was prepared by dissolving the RNA in an aqueous solution (10 mM sodium phosphate buffer, pH 6.7, 20 mM NaCl, 95% H2O, 5% 2H2O or 100% 2H2O), heated to 368 K and snap-cooled onto ice. The sample concentration was 2.0 mM. The sample used to measure RDCs was prepared (1.0 mM in RNA) by mixing the oligonucleotide with a 50 mg/ml filamentous Pf1 phage solution (Asla Ltd) to a total phage concentration of 17 mg/ml (16). Deuterium splitting was 15.2 Hz at 800 MHz. UV melting of the construct was measured using a Perkin–Elmer Lambda Bio 40 spectrometer equipped with a Peltier system. The melting temperature was 343 K (1.9 μM oligonucleotide, 10 mM sodium phosphate buffer, pH 6.7, 20 mM NaCl).
NMR spectroscopy
NMR data were acquired on Varian NMR Systems 600 and 800 MHz NMR spectrometers. Spectra were processed and analyzed using VNMRJ 2.1B (Varian Inc.) and Felix 2002 software (Accelrys Inc.). 1H, 13C and 15N resonances were assigned from a combined analysis of 2D homonuclear experiments (NOESY, TOCSY, DQF-COSY), 2D 13C/15N-HSQC, 2D nucleobase-specific experiments (HCCH-TOCSY, HNCCCH and HNC-TOCSY-CH), 2D HNN-COSY, 2D H(N)CO, 2D HCN and 3D NOESY-13C-HSQC (17–23). One-bond 1H-15N and 1H-13C RDCs were determined by simulation of F1 doublets (error ± 2.0 Hz) in 2D IPAP 15N-HSQC (24) and 2D CE-CT 13C-HSQC spectra (25), respectively.
Restraints and structure calculations
2D NOESY spectra recorded at 298 K with mixing times of 75, 150 and 300 ms for the 100% 2H2O sample were used to obtain NOE distance restraints for nonexchangeable protons with the upper and lower bounds set to ±20%, ±30% and ±40%, respectively. The volume of the pyrimidine H5-H6 cross-peak was used to set the reference distance of 2.45 Å. Other proton–proton distances were calculated using the I ∼ rij –6 relation. NOE distance restraints of exchangeable protons were obtained from the 2D NOESY spectra of the 95% H2O sample recorded with a 75 and 300 ms mixing time at 278 and 298 K with WATERGATE pulse sequence for solvent suppression. Cross-peaks were classified as strong (1.8–3.6 Å), medium (2.6–5.0 Å) and weak (3.5–6.5 Å). Additional NOE distance restraints for nonexchangeable protons were obtained from two 3D NOESY-13C-HSQC spectra (one for aromatic and one for ribose protons) recorded at 298 K with 150 ms mixing time. Altogether, 693 NOE distance restraints were used in the structure calculation, out of which 74 were from exchangeable protons. The δ torsion angle was restrained to N-type sugar geometry for nucleotides with absent H1′-H2′ cross-peaks in the DQF-COSY spectrum. Torsion angles ν0–ν4 of U24 were restrained to the values characteristic for S-type sugar geometry. The χ torsion angles of nucleotides in the stems were estimated from the intensities of intranucleotide H6/H8-H1′/H2′ cross-peaks in 2D NOESY spectra, and were therefore set to anti (210 ± 40°). The β torsion angles of the stem nucleotides were restrained to trans (180 ± 40°) due to the absence of P-H5′ and P-H5″ cross-peaks in the 2D HP-COSY spectrum. The ε torsion angle was restrained to trans for all stem nucleotides. Torsion angles α and ζ were set to exclude the trans conformation due to the narrow range of 31P resonances. We did not apply any backbone torsion restraints for residues in the asymmetric internal loop and GAAA tetraloop. Experimental RDC values were used in the simulations directly.
Structure calculations were performed using AMBER 9 (26) with a Wang et al. (27) force field. Initial starting structures were created using 8 ps unrestrained MD at different temperatures from 300 to 2000 K. Structures were then subjected to 60 ps of restrained simulated annealing (SA) calculations using a generalized Born implicit solvation model. The molecules were heated to 1000 K during the first 5 ps, after which temperature was constant for 30 ps, scaled down to 100 K in the next 11 ps and reduced to 0 K in the last 14 ps. The force constants were 35 kcal mol−1 Å−2 for NOE distance, 300 kcal mol−1 rad−2 for torsion angle and 25 kcal mol−1 Å−2 for base planarity restraints. The cutoff for nonbonded interactions was 20 Å. The SHAKE algorithm for hydrogen atoms was used with a tolerance of 0.0005 Å. All structures from SA were subjected to a maximum of 10 000 steps of conjugate gradient minimization. A family of 10 minimized structures with the lowest energy and the smallest NMR violations were analyzed using ptraj and suppose programs. Helical parameters for the two stems were analyzed with the computer program 3DNA (28).
Coordinate deposition
The coordinates for the family of the lowest energy structures were deposited in the Protein Data Bank with accession code 2JXV.
RESULTS
The 33-nt construct folds into a well-defined structure
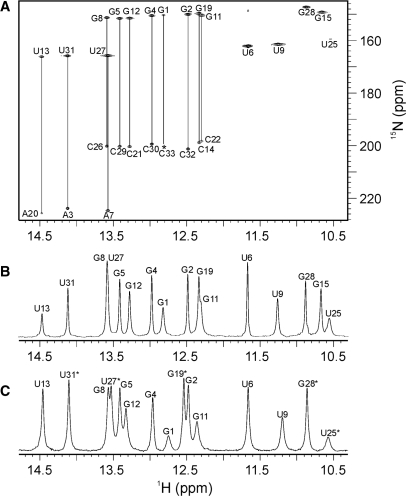
Following the isolation and purification steps, the sample of the 33-nt RNA oligonucleotide was transferred into an NMR tube. The 1D proton NMR spectrum displays nine well-resolved peaks in the region from 12.3 to 14.5 p.p.m., which is typical for Watson–Crick imino proton resonances (Figure 2B). The five additional imino proton resonances were found in the region from 10.5 to 11.7 p.p.m. and were assigned to non Watson–Crick base-paired residues from the asymmetric internal loop, the GAAA tetraloop and the GU wobble base pair. Base pairing network was initially analyzed through imino–imino and imino–amino sequential walks in 2D NOESY spectra, and subsequently confirmed by the HNN-COSY spectrum, which correlated each imino proton with two nitrogen atoms within the regular GC and AU Watson–Crick base pairs. The most upfield cross-peaks with the nitrogen chemical shifts in the range from 146 to 152 p.p.m. belong to ten guanine N1 atoms (Figure 2A). Eight of them show correlations with the corresponding cytosine N3 atoms with the characteristic chemical shifts in the range from 197 to 202 p.p.m. The cross-peaks with nitrogen resonances in the chemical shift range from 159 to 167 p.p.m. belong to N3 atoms of six uracils. U13, U27 and U31 are correlated with Watson–Crick hydrogen-bonded partners, A20, A7 and A3, respectively, which exhibit N1 chemical shifts in the range from 223 to 226 p.p.m. (Figure 2A). Imino protons of U6, U9, G15, U25 and G28 do not show correlations with adenine or cytosine 15N atoms, which suggests that they are not involved in Watson–Crick base pairs.
Figure 2.
(A) HNN-COSY spectrum of 33-nt RNA molecule showing A-U and G-C base pairs (indicated by vertical lines). A single cross-peak at a given 1H chemical shift indicates residue not involved in Watson–Crick base pair. Asterisk denotes the cross-peak of C33 with low intensity. (B) Imino proton region of 1D NMR spectrum of 33-nt RNA molecule. (C) Imino proton region of 1D NMR spectrum of the dimeric construct.
Cytosine amino groups have been assigned through characteristic correlations in a 2D 15N-HSQC spectrum. Each of the eight amino nitrogen resonances (δ 100–104 p.p.m.) exhibit correlations with two proton resonances (data not shown). The downfield proton resonances correspond to hydrogen-bonded amino protons (δ 8.0–8.5 p.p.m.), while the corresponding upfield resonances belong to the protons not involved in hydrogen bonds (δ 6.6–7.3 p.p.m.).
The assignment of the three AU and eight GC Watson–Crick base pairs has enabled us to establish the secondary structure of the RNA construct in which the asymmetric internal loop separates the lower (residues 1–8 and 26–33) and upper stems (residues 11–14 and 19–22), while the GAAA tetraloop connects the two antiparallel strands (Figure 1B). The lower stem is stabilized by the 8 bp with U6·G28 being the wobble base pair. Three GC and one AU base pair stabilize the upper stem. The chemical shifts of U9, G15 and U25 imino protons are indicative of noncanonical base pairs which contribute to the stabilization of the internal and apical loops. The imino proton of U24 could not be observed neither at 278 K nor 298 K presumably due to its exposure to fast exchange with bulk solvent. No cross-peak was observed in the HNCCCH spectrum for U24, whereas imino protons of U9 and U25 showed correlations to their H6 protons (see Figure S1 in the Supplementary Data).
Resonance assignment
Our assignment procedure could be dissected into three steps. One of the steps established mutual intranucleotide correlations between imino, amino and aromatic protons and heteroatoms with the use of homonuclear and heteronuclear base experiments. H6 of cytosine and uracil residues were assigned through their correlations with amino and imino protons. H5 protons were assigned with the use of a 2D TOCSY spectrum which exhibited resolved correlations between H5 (δ 5.0–5.9 p.p.m.) and H6 proton resonances (δ 7.4–8.1 p.p.m.). The assignment of guanine H8 proton resonances was done through their correlations with imino protons within a guanine base. H2 protons of A3, A7 and A20 showed resolved correlations with uracil imino protons in the 2D NOESY spectrum. In addition, H2 and H8 protons of adenine residues, including those not involved in AU base pairs, were correlated with the use of a 2D HCCH-TOCSY spectrum.
The next step in our assignment procedure led to the identification of the ribose protons. 3D HCCH-COSY, 3D HCCH-RELAY and 3D HCCH-TOCSY spectra helped us to perform the assignment of all ribose protons including H5′ and H5″. We noticed some unusual chemical shifts which were, however, characteristic for a GAAA tetraloop. For instance, the chemical shift of H3′ of A18, the last adenine in the tetraloop, appeared downfield at 5.13 p.p.m. H1′ of G19 which follows the GAAA tetraloop at the 3′ end resonates at 3.61 p.p.m. We noticed that ribose cross-peaks of G15, A16 and A17 were weak which suggested that their sugar moieties were involved in an unbiased pseudorotational equilibrium.
In the last step of our assignment, aromatic and ribose spin systems were coupled together using a 2D HCN experiment, which correlated most of the H1′ and H8 to purine N9, and H1′ and H6 to pyrimidine N1.
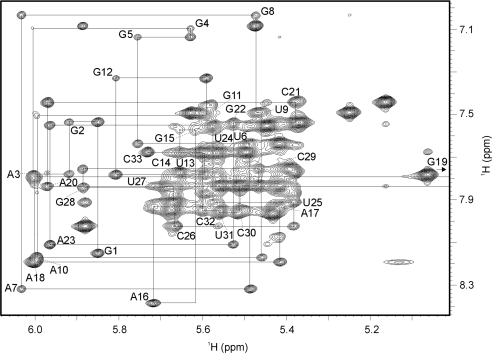
The problem of considerable spectral overlap in the ribose region of 2D NOESY spectra was reduced by utilizing 3D NOESY-13C-HSQC spectra. As an example, sequential correlations using the strips from these spectra for residues 6–12 and 21–28 are shown in the Supplementary Data (Figure S2). Assignment of all protons allowed us to do the complete sequential walk between H6/H8 and H1′ proton resonances in the 2D NOESY spectrum (Figure 3). Weak sequential NOE cross-peaks were observed between C14 H1′ and G15 H8, and between G15 H1′ and A16 H8 which are due to the long distances imposed by stacking of A16, A17 and A18 bases in the GAAA tetraloop. Intranucleotide H6/H8-H1′ cross-peaks displayed characteristic intensities of nucleobases in the anti conformation for all residues.
Figure 3.
The aromatic-anomeric region of 2D NOESY spectrum (τm = 300 ms). Lines indicate the sequential walk.
The 2D HP-COSY and 3D HCP spectra enabled us to assign 27 out of 33 phosphorus resonances. The chemical shifts for most phosphorus atoms were within −0.9 and 0.2 p.p.m. The 31P resonances of A16, A17, A18 and G19 were found in the range from −1.1 to 1.8 p.p.m. A16 exhibited the most downfield signal (δ 1.8 p.p.m), whereas 31P of A18 was the most upfield (δ −1.1 p.p.m.), which was as expected for the GAAA tetraloop (29).
Qualitative structural information on the dimeric construct
In response to the comments of anonymous referee we have analyzed a dimeric construct consisting of two separate strands that originate from lin-41 mRNA and let-7 miRNA (Figure 1C). 1D 1H spectrum of this dimer shows high degree of similarity with the 33-nt construct (cf. Figure 2B and C). 2D NOESY spectrum of the dimeric construct is available in the Supplementary Data (Figure S3). We could perform a complete H8-H1′ sequential walk with the help of the chemical shift data for the 33-nt stem–loop construct. Both 1D and NOESY spectra of stem–loop and dimer constructs show evident similarities, and are almost identical except for the GAAA tetraloop which is replaced with the closing GC base pair in the latter.
Residual dipolar couplings
One-bond 1H-15N residual dipolar couplings (RDCs) were measured from the splitting of the peaks along the nitrogen dimension of the 2D IPAP 15N-HSQC spectrum and comparing the data for the samples before and after addition of Pf1 phage (Figure S4). The spectra used for quantification of RDC values were of high quality with narrow line widths showing no apparent signs of aggregation or interaction between phage and RNA. Altogether, 14 one-bond 1H-15N RDC values were collected with values in the range from −17.7 to −5.0 Hz (Table S1). The lower stem exhibited one-bond 1H-15N RDC values between −17.7 and −11.7 Hz and the upper stem between −11.5 and −10.7 Hz. These RDC values thus implied that the upper stem was not collinear with the lower stem. One-bond 1H-13C RDC values were measured for H2-C2, H6-C6 and H8-C8 bonds. These RDC values were found in the range from 8.3 to 34.3 Hz and were similar for the lower and upper stems. Residues G4, G5 and A7 from the lower stem exhibited higher 1H-13C RDC values than the other residues. G1 and C33 exhibited the lowest values which were attributed to the end frying. GAAA tetraloop residues exhibited 1H-13C RDC values in the range from 12.1 to 20.1 Hz, while for residues in the asymmetric internal loop the corresponding values were in the range from 22.3 to 34.7 Hz.
Sugar conformation
The 2D DQF-COSY spectrum indicated the absence of most of H1′-H2′ cross-peaks, which suggested that sugars in the stem regions adopted predominantly an N-type conformation. However, U24 exhibited a high 3J1’2’ coupling constant (∼8 Hz) implying the predominance of S-type conformation for its sugar moiety. We observed weak H1′-H2′ cross-peaks for residues A16, A17, A23 and C33, which indicated that their sugar rings are involved in an unbiased N-S conformational equilibrium. Consequently, sugar conformations of these four residues were not restrained in the course of structure calculations.
Structural restraints
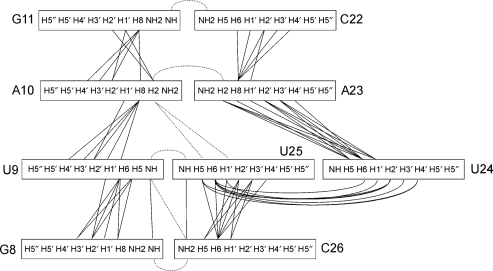
NOE cross-peak volumes were translated into distances using the Felix 2002 program (Accelrys Inc.). 2D NOESY spectra acquired in 95% H2O were used for distance calculations involving exchangeable protons (imino–imino, imino–amino and imino–H2 cross-peaks). These types of distance restraints were particularly valuable to establish spatial relations of sequential nucleotides as well as connectivities across the asymmetric internal loop. The distances between nonexchangeable protons were derived from 2D NOESY spectra acquired in 100% 2H2O. Due to the considerable overlap in 2D NOESY spectra only resolved cross-peaks were used in the calculation of distance restraints. The 3D NOESY-13C-HSQC spectra were then used to quantitate the volumes of additional cross-peaks which were classified as strong, medium and weak. Internucleotide NOE contacts in the asymmetric internal loop are displayed schematically in Figure 4. We noticed that the asymmetric internal loop exhibited a smaller number of NOEs per residue than the rest of the molecule.
Figure 4.
Schematic presentation of internucleotide NOE contacts in the asymmetric internal loop and the closing GC base pairs. Sequential NOE connectivities are shown with a full line, whereas long-range NOE contacts are shown as dotted lines.
Structure determination
693 NOE-derived distance restraints (on average 21 NOE restraints per residue), 130 torsion angle restraints and 51 RDC values were used in the process of structure determination (Table 1). Thirty-two hydrogen bond and 22-bp planarity restraints were applied for GC, AU and GU base pairs in the two stems. No hydrogen bond and planarity restraints were used within the asymmetric internal and terminal loops. After extensive simulated annealing and energy minimization steps on over 100 structures our calculations converged to a family of 10 structures that agreed with all the available NMR data. NOE distance violations for the final set of 10 structures were below 0.4 Å and torsion angle violations were below 5°. A perusal of the family of final structures shows that all parts of the 33-nt construct are well-defined (Figure 5). Residues U9, U24 and U25 in the asymmetric internal loop form a base triple, while A10 and A23 form a base pair (Figure 6). The structure of the GAAA tetraloop is similar to structures described in literature (30,31). Residues G15 and A18 form a sheared GA mismatch, while A16, A17 and A18 are stacked with each other.
Table 1.
NMR restraints and structural statistics
| Distance restraints | |
| Intranucleotide NOEs | 370 |
| Internucleotide NOEs (n, n + 1) | 266 |
| Long-range NOEs (n, n + m, m > 1) | 57 |
| Torsion angle restraints | 130 |
| Hydrogen bond restraints | 32 |
| Residual dipolar couplings | 51 |
| Base pair planarity restraints | 22 |
| Total number of restraints | 928 (∼28/nt) |
| NOE violations > 0.3 Å | 2.2 ± 1.2 |
| Maximum NOE violations (Å) | 0.38 ± 0.08 |
| Deviations from idealized covalent geometry | |
| Bonds (Å) | 0.009 ± 0.000 |
| Angles (°) | 1.62 ± 0.12 |
| Pairwise all heavy atom RMSD from average structure (Å) | |
| Overall | 0.76 ± 0.20 |
| Without the asymmetric internal loop | 0.76 ± 0.24 |
| Without the GAAA tetraloop | 0.63 ± 0.29 |
| Without the first GC base pair | 0.74 ± 0.22 |
| Without the GAAA tetraloop and the first GC base pair | 0.62 ± 0.31 |
| Without the asymmetric internal loop and the GAAA tetraloop | 0.64 ± 0.33 |
| Without the asymmetric internal loop, the GAAA tetraloop and the first GC base pair | 0.62 ± 0.36 |
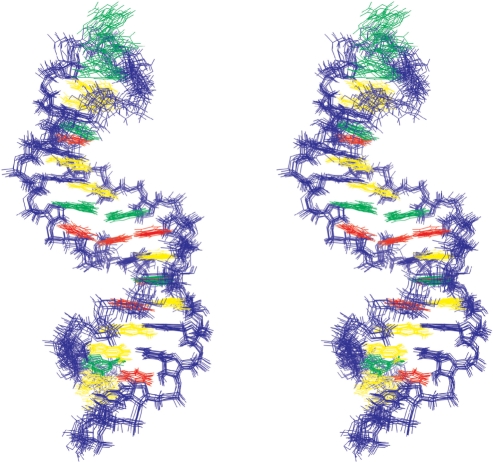
Figure 5.
Stereo view of the family of 10 lowest energy structures. Sugar-phosphate backbone and cytosine bases are colored in blue, adenine bases in green, uracil bases in red and guanine bases in yellow.
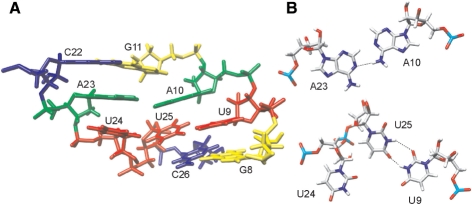
Figure 6.
Structural details within the asymmetric internal loop. (A) Side view of the average structure. Cytosines are colored in blue, adenines in green, uracils in red and guanines in yellow. (B) A10·A23 base pair and U9·U25·U24 base triple shown from the top. Dashed lines indicate hydrogen bonds.
Overall pairwise heavy atom RMSD to the mean was 0.76 Å for the 10 superimposed structures. The RMSD within the family of final structures without considering residues of the asymmetric internal loop was similar. The comparison of structures without taking into account the GAAA tetraloop and the first GC base pair gave a lower RMSD value of 0.62 Å (Table 1).
Simulated annealing calculations proposed two more possible structures for the asymmetric internal loop. One structure exhibited nucleotide U24 positioned coplanar with the G8-C26 base pair, whereas the other one exhibited U24 in a flipped out conformation. However, both of these structures were energetically less favorable in comparison to those shown in Figure 5.
DISCUSSION
mRNAs were for a long time viewed as simple strings of codons programmed to carry genetic information from the DNA template to the ribosome. It has been recently shown that mRNA can be targeted to control gene expression. RNA interference, chosen as the breakthrough of the year 2002 by the Science magazine (32), describes the ability of small and apparently structurally simple RNA molecules to switch off or regulate expression of specific genes in a wide variety of organisms ranging from plants to humans by binding to stretches of mRNA. Insight into the intricate mechanisms that induce the silencing function of the short RNAs is of critical importance in understanding their biological roles as well as in potential applications as drugs.
The 33-nt RNA construct, which mimics a complex between the C. elegans let-7 miRNA and its imperfectly complementary site from the 3'-UTR of the lin-41 mRNA (LCS 2), has been shown to fold into a well-defined stem–loop structure (Figure 5). NMR data and subsequent structure calculations have demonstrated that the structure exhibits two stems which are separated by an asymmetric internal loop. The lower stem is stabilized by seven Watson–Crick and a GU wobble base pair and shows conformational features of A-form RNA. Hydrogen bonds within the U6·G28 base pair are formed between the U6 imino proton and the G28 O6 atom, and between the G28 imino proton and the U6 O2 atom (33). The amino group of G28 points into the minor groove. The two bases of the GU base pair are rotated differently in comparison to the standard AU and GC Watson–Crick base pairs. The λ angle between the glycosidic bond and C1′-C1′ line in a base pair is 69° for U6 and 38° for G28, while it is ∼54° for residues involved in Watson–Crick base pairs. In addition, the U6·G28 wobble base pair affects the helical twist (∼41°) with the A7·U27 base pair. The upper stem consists of four Watson–Crick base pairs and exhibits an average twist and rise parameters of 32.7° and 3.0 Å, respectively. For comparison, twist and rise parameters in A-form RNA are 32.7° and 2.8 Å, respectively (34).
The asymmetric internal loop adopts a single well-defined structure (Figure 6A). U9 and U25 form a base pair with hydrogen bonds between imino protons and carbonyl groups (Figures 6B and S5) similar to the UU base pair described earlier (35). The residue U24 is placed in a coplanar arrangement with respect to U9 and U25 thus forming a base triple (Figure 6B). The puckering of the sugar ring of U24 is in the South conformation (C2′-endo) with the phase angle of pseudorotation and the maximum puckering amplitude equal to 162° and 24°, respectively. In order to position nucleotide U24 next to U25, the torsion angles ζ in U24 and α, β and ε in U25 adopt conformations outside their usual ranges found in A-form RNA. A10 and A23 form a base pair (36) with a hydrogen bond between the A10 amino proton and A23 N1 (Figure 6B). The A10·A23 base pair is not corroborated by the observable amino group protons, which could have been resolved from a crowded amino–amino region of the NOESY spectra. However, this base pair is supported by the correlation peak between the A10 H2 and A23 H2 proton observed in the 3D NOESY-13C-HSQC spectrum. The stacking of the nucleobases in the asymmetric internal loop is extensive and probably contributes to a well-defined and thermodynamically stable structure. The slide of the A10·A23 with respect to the U9·U25 base pair positions A10 above the U9·U25 base pair, and A23 above U24 (Figure 6A). The G8-C26 and G11-C22 base pairs close the asymmetric internal loop at the two ends. The U9·U25 base pair is stacked over the G8-C26 base pair. G11 is stacked over the center of the A10·A23 base pair, and C22 is stacked on A23. The helical twist between the G8-C26 and U9·U25 base pairs is around 36°.
The GAAA tetraloop exhibits characteristic and expected chemical shifts and structural features (31). NMR spectra clearly show that the G15 imino proton is not freely accessible to solvent. The tetraloop thus consists of a sheared G15·A18 mismatch base pair and A16 and A17 which stack on A18. The G15·A18 base pair has a small helical twist (∼10°) which positions A18 directly above G19 H1'. Additional hydrogen bonds, e.g. between the G15 amino proton and phosphate oxygen in the A16pA17 step, make the tetraloop very stable. Sugar puckers of adenine nucleotides in the GAAA tetraloop display unbiased pseudorotational equilibrium between N- and S-puckered conformations.
One-bond 1H-15N and 1H-13C RDC values suggest that the lower and upper stems are not completely collinear. The analysis of helical parameters of the lower and upper stems showed that their helical axes exhibited an angular bend of ∼20°. In recent years, several RNA structures have been determined which consist of internal loops, bulges or noncanonical base pairs (37–43). Leeper and Varani (37) have shown that the (2 + 3) internal loop ‘J6’ in the structure of an enzyme-activating fragment of the human telomerase RNA introduces an angular bend of 20° between the two helical axes. This value is similar to the angular bend found in our construct. The structures of the two asymmetric internal loops show some similarities, although consisting of different nucleotides. For example, the base-stacking pattern between C288, A289 and C290 in Varani's structure is analogous to the base-stacking between C22, A23 and U24 in our construct. A potential C266·U291 base pair in the former is similar to the U9·U25 base pair in the construct studied here. The two internal loops differ, however, in base triple formation. C267 is hypothesized to lie coplanar with the G268-C288 base pair outside the internal loop (37). U24, which is in our model system on the opposite side of the asymmetric internal loop, is coplanar with U9 and U25. A10 and A23 form a base pair and stack with G11-C22 base pair which closes the asymmetric internal loop.
The solution structure of the apical stem–loop of the human hepatitis B virus encapsidation signal shows that a single nucleotide bulge induces ∼20° bend between the lower and the upper helices, which leads to an increase in deepness of the major groove (39). The asymmetric internal loop in our model system bends both stems in such a way that the major groove becomes wider.
Asymmetric internal loops and mismatch base pairs are common for the miRNA:mRNA complexes. Several studies have shown that base pairing, asymmetric internal loops, and the bulged A residues and GU wobble base pairs between let-7 and lin-41 are critical for downregulation of lin-41 (44,45). Mutations in the anticipated asymmetric internal loops, the lower and upper stems in let-7 complementary sites dramatically influence miRNA silencing of the target genes. The 3D structure of our the 33-nt construct mimicking the let-7:lin-41 complex gives new structural insights as to why certain mutations are not tolerated and are thus more important for function than others.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
The authors would like to thank Professor Gabriele Varani (University of Washington) for helpful information on RNA synthesis. Financial support from the Slovenian Research Agency (ARRS) and the Ministry of Higher Education, Science and Technology of the Republic of Slovenia (Grant No. P1-0242-0104) is gratefully acknowledged. Funding to pay the Open Access publication charges for this article was provided by ARRS.
Conflict of interest statement. None declared.
REFERENCES
- 1.Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, Barzilai A, Einat P, Einav U, Meiri E, et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat. Genet. 2005;37:766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 3.Niwa R, Slack FJ. The evolution of animal microRNA function. Curr. Opin. Genet. Dev. 2007;17:145–150. doi: 10.1016/j.gde.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 4.MacRae IJ, Zhou K, Doudna JA. Structural determinants of RNA recognition and cleavage by Dicer. Nat. Struct. Mol. Biol. 2007;14:934–940. doi: 10.1038/nsmb1293. [DOI] [PubMed] [Google Scholar]
- 5.Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 7.Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Muller P, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 8.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 9.Schulman BRM, Esquela-Kerscher A, Slack FJ. Reciprocal expression of lin-41 and the microRNAs let-7 and mir-125 during mouse embryogenesis. Dev. Dyn. 2005;234:1046–1054. doi: 10.1002/dvdy.20599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat. Rev. Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 11.Johnson CD, Esquela-Kerscher A, Stefani G, Byrom M, Kelnar K, Ovcharenko D, Wilson M, Wang X, Shelton J, Shingara J, et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 2007;67:7713–7722. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- 12.Lingel A, Simon B, Izaurralde E, Sattler M. Nucleic acid 3′-end recognition by the Argonaute2 PAZ domain. Nat. Struct. Mol. Biol. 2004;11:576–577. doi: 10.1038/nsmb777. [DOI] [PubMed] [Google Scholar]
- 13.Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, Eachus R, Pasquinelli AE. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 14.Milligan JF, Uhlenbeck OC. Synthesis of small RNAs using T7 RNA polymerase. Method Enzymol. 1989;180:51–62. doi: 10.1016/0076-6879(89)80091-6. [DOI] [PubMed] [Google Scholar]
- 15.Kao C, Zheng M, Rudisser S. A simple and efficient method to reduce nontemplated nucleotide addition at the 3′ terminus of RNAs transcribed by T7 RNA polymerase. RNA. 1999;5:1268–1272. doi: 10.1017/s1355838299991033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hansen MR, Mueller L, Pardi A. Tunable alignment of macromolecules by filamentous phage yields dipolar coupling interactions. Nat. Struct. Biol. 1998;5:1065–1074. doi: 10.1038/4176. [DOI] [PubMed] [Google Scholar]
- 17.Nikonowicz EP, Pardi A. An efficient procedure for assignment of the proton, carbon and nitrogen resonances in 13C/15N labeled nucleic acids. J. Mol. Biol. 1993;232:1141–1156. doi: 10.1006/jmbi.1993.1466. [DOI] [PubMed] [Google Scholar]
- 18.Marino JP, Prestegard JH, Crothers DM. Correlation of adenine H2/H8 resonances in uniformly 13C labeled RNAs by 2D HCCH-TOCSY: a new tool for 1H assignment. J. Am. Chem. Soc. 1994;116:2205–2206. [Google Scholar]
- 19.Simorre JP, Zimmermann GR, Pardi A, Farmer BTII, Mueller L. Triple resonance HNCCCH experiments for correlating exchangeable and nonexchangeable cytidine and uridine base protons in RNA. J. Biomol. NMR. 1995;6:427–432. doi: 10.1007/BF00197641. [DOI] [PubMed] [Google Scholar]
- 20.Simorre JP, Zimmermann GR, Mueller L, Pardi A. Correlation of the guanosine exchangeable and nonexchangeable base protons in 13C-/15N-labeled RNA with an HNC-TOCSY-CH experiment. J. Biomol. NMR. 1996;7:153–156. doi: 10.1007/BF00203825. [DOI] [PubMed] [Google Scholar]
- 21.Wijmenga SS, van Buuren BNM. The use of NMR methods for conformational studies of nucleic acids. Prog. Nucl. Magn. Reson. Spectrosc. 1998;32:287–387. [Google Scholar]
- 22.Dingley AJ, Grzesiek S. Direct observation of hydrogen bonds in nucleic acid base pairs by internucleotide 2JNN couplings. J. Am. Chem. Soc. 1998;120:8293–8297. [Google Scholar]
- 23.Furtig B, Richter C, Wohnert J, Schwalbe H. NMR spectroscopy of RNA. ChemBioChem. 2003;4:936–962. doi: 10.1002/cbic.200300700. [DOI] [PubMed] [Google Scholar]
- 24.Ottiger M, Delaglio F, Bax A. Measurement of J and dipolar couplings from simplified two-dimensional NMR spectra. J. Magn. Reson. 1998;131:373–378. doi: 10.1006/jmre.1998.1361. [DOI] [PubMed] [Google Scholar]
- 25.Tian F, Al-Hashimi HM, Craighead JL, Prestegard JH. Conformational analysis of a flexible oligosaccharide using residual dipolar couplings. J. Am. Chem. Soc. 2001;123:485–492. doi: 10.1021/ja002900l. [DOI] [PubMed] [Google Scholar]
- 26.Case DA, Darden T, Cheatham TEIII, Simmerling C, Wang J, Duke RE, Luo R, Merz KM, Pearlman DA, Crowley M, et al. San Francisco, CA: University of California; 2006. AMBER 9. [Google Scholar]
- 27.Wang JM, Cieplak P, Kollman PA. How well does a restrained electrostatic potential (RESP) model perform in calculating conformational energies of organic and biological molecules? J. Comput. Chem. 2000;21:1049–1074. [Google Scholar]
- 28.Lu XJ, Olson WK. 3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 2003;31:5108–5121. doi: 10.1093/nar/gkg680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Legault P, Pardi A. 31P chemical shift as a probe of structural motifs in RNA. J. Magn. Reson. Ser. B. 1994;103:82–86. doi: 10.1006/jmrb.1994.1012. [DOI] [PubMed] [Google Scholar]
- 30.Heus HA, Pardi A. Structural features that give rise to the unusual stability of RNA hairpins containing GNRA loops. Science. 1991;253:191–194. doi: 10.1126/science.1712983. [DOI] [PubMed] [Google Scholar]
- 31.Jucker FM, Heus HA, Yip PF, Moors EHM, Pardi A. A network of heterogeneous hydrogen bonds in GNRA tetraloops. J. Mol. Biol. 1996;264:968–980. doi: 10.1006/jmbi.1996.0690. [DOI] [PubMed] [Google Scholar]
- 32.Couzin J. Breakthrough of the year: small RNAs make big splash. Science. 2002;298:2296–2297. doi: 10.1126/science.298.5602.2296. [DOI] [PubMed] [Google Scholar]
- 33.Varani G, McClain WH. The GU wobble base pair - a fundamental building block of RNA structure crucial to RNA function in diverse biological systems. EMBO Rep. 2000;1:18–23. doi: 10.1093/embo-reports/kvd001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saenger W. Princliples of Nucleic Acid Structure. 1984. Springer, New York, NY, pp. 242–252. [Google Scholar]
- 35.Lietzke SE, Barnes CL, Berglund JA, Kundrot CE. The structure of an RNA dodecamer shows how tandem U-U base pairs increase the range of stable RNA structures and the diversity of recognition sites. Structure. 1996;4:917–930. doi: 10.1016/s0969-2126(96)00099-8. [DOI] [PubMed] [Google Scholar]
- 36.Fourmy D, Yoshizawa S, Puglisi JD. Paromomycin binding induces a local conformational change in the A-site of 16 S rRNA. J. Mol. Biol. 1998;277:333–345. doi: 10.1006/jmbi.1997.1551. [DOI] [PubMed] [Google Scholar]
- 37.Leeper TC, Varani G. The structure of an enzyme-activating fragment of human telomerase RNA. RNA. 2005;11:394–403. doi: 10.1261/rna.7222505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Theimer CA, Feigon J. Structure and function of telomerase RNA. Curr. Opin. Struct. Biol. 2006;16:307–318. doi: 10.1016/j.sbi.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 39.Flodell S, Petersen M, Girard F, Zdunek J, Kidd-Ljunggren K, Schleucher J, Wijmenga S. Solution structure of the apical stem-loop of the human hepatitis B virus encapsidation signal. Nucleic Acids Res. 2006;34:4449–4457. doi: 10.1093/nar/gkl582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Erat MC, Zerbe O, Fox T, Sigel RKO. Solution structure of domain 6 from a self-splicing group II intron ribozyme: a Mg2+ binding site is located close to the stacked branch adenosine. ChemBioChem. 2007;8:306–314. doi: 10.1002/cbic.200600459. [DOI] [PubMed] [Google Scholar]
- 41.Zoll J, Tessari M, Van Kuppeveld FJM, Melchers WJG, Heus HA. Breaking pseudo-twofold symmetry in the poliovirus 3′-UTR Y-stem by restoring Watson-Crick base pairs. RNA. 2007;13:781–792. doi: 10.1261/rna.375607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shankar N, Xia T, Kennedy SD, Krugh TR, Mathews DH, Turner DH. NMR reveals the absence of hydrogen bonding in adjacent UU and AG mismatches in an isolated internal loop from ribosomal RNA. Biochemistry. 2007;46:12665–12678. doi: 10.1021/bi700802s. [DOI] [PubMed] [Google Scholar]
- 43.Sashital DG, Venditti V, Angers CG, Cornilescu G, Butcher SE. Structure and thermodynamics of a conserved U2 snRNA domain from yeast and human. RNA. 2007;13:328–338. doi: 10.1261/rna.418407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vella MC, Reinert K, Slack FJ. Architecture of a validated microRNA::target interaction. Chem. Biol. 2004;11:1619–1623. doi: 10.1016/j.chembiol.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 45.Vella MC, Choi EY, Lin SY, Reinert K, Slack FJ. The C. elegans microRNA let-7 binds to imperfect let-7 complementary sites from the lin-41 3′ UTR. Genes Dev. 2004;18:132–137. doi: 10.1101/gad.1165404. [DOI] [PMC free article] [PubMed] [Google Scholar]