Abstract
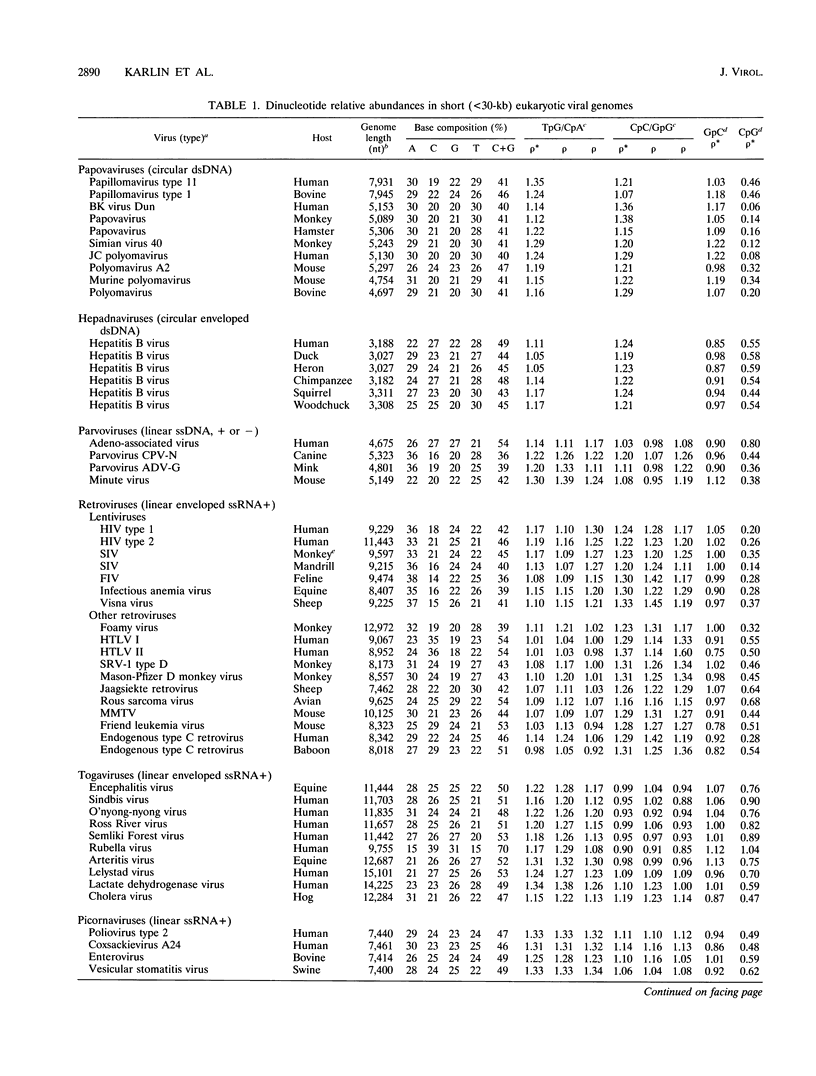
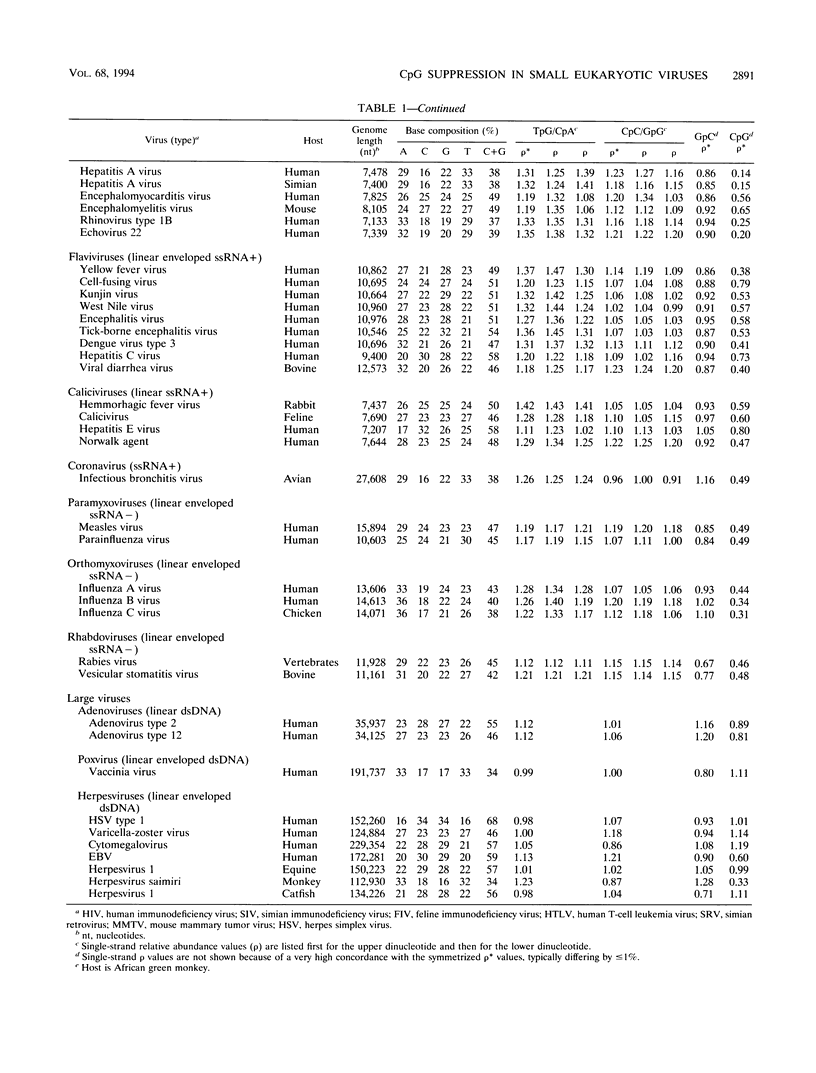
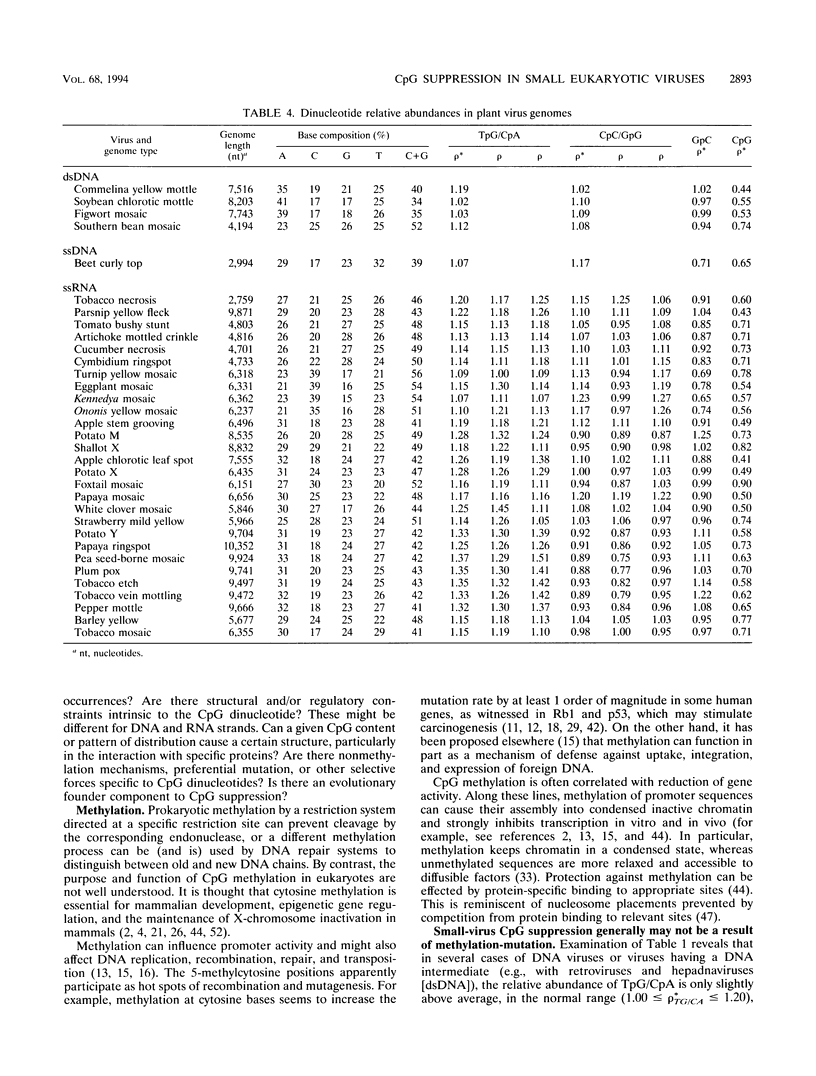
Dinucleotide over- and underrepresentation is evaluated in all available completely sequenced DNA or RNA viral genomes, ranging in size from 3 to 250 kb (available RNA viruses fall into the small-virus category). The dinucleotide CpG is statistically underrepresented (suppressed) in all but four of the small viruses (more than 75 with lengths of < 30 kb) but has normal relative abundances in most large viruses (> or = 30 kb). Most retrotransposons in eukaryotic species also show low CpG relative abundances. Interpretations, especially in some cases of DNA viruses or viruses with a DNA intermediate, might relate to methylation effects and modes of viral integration and excision. Other possible contributing factors relate to dinucleotide stacking energies, special mutation mechanisms, and evolutionary events.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berns K. I., Labow M. A. Parvovirus gene regulation. J Gen Virol. 1987 Mar;68(Pt 3):601–614. doi: 10.1099/0022-1317-68-3-601. [DOI] [PubMed] [Google Scholar]
- Bestor T. H., Coxon A. Cytosine methylation: the pros and cons of DNA methylation. Curr Biol. 1993 Jun 1;3(6):384–386. doi: 10.1016/0960-9822(93)90209-7. [DOI] [PubMed] [Google Scholar]
- Beutler E., Gelbart T., Han J. H., Koziol J. A., Beutler B. Evolution of the genome and the genetic code: selection at the dinucleotide level by methylation and polyribonucleotide cleavage. Proc Natl Acad Sci U S A. 1989 Jan;86(1):192–196. doi: 10.1073/pnas.86.1.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. P. Genomic imprinting: imprints on islands. Curr Biol. 1993 May 1;3(5):275–277. doi: 10.1016/0960-9822(93)90177-p. [DOI] [PubMed] [Google Scholar]
- Birkenbach M., Josefsen K., Yalamanchili R., Lenoir G., Kieff E. Epstein-Barr virus-induced genes: first lymphocyte-specific G protein-coupled peptide receptors. J Virol. 1993 Apr;67(4):2209–2220. doi: 10.1128/jvi.67.4.2209-2220.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge C., Campbell A. M., Karlin S. Over- and under-representation of short oligonucleotides in DNA sequences. Proc Natl Acad Sci U S A. 1992 Feb 15;89(4):1358–1362. doi: 10.1073/pnas.89.4.1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challberg M. D., Kelly T. J. Animal virus DNA replication. Annu Rev Biochem. 1989;58:671–717. doi: 10.1146/annurev.bi.58.070189.003323. [DOI] [PubMed] [Google Scholar]
- Cooper D. N. Eukaryotic DNA methylation. Hum Genet. 1983;64(4):315–333. doi: 10.1007/BF00292363. [DOI] [PubMed] [Google Scholar]
- Cooper D. N., Youssoufian H. The CpG dinucleotide and human genetic disease. Hum Genet. 1988 Feb;78(2):151–155. doi: 10.1007/BF00278187. [DOI] [PubMed] [Google Scholar]
- Delcourt S. G., Blake R. D. Stacking energies in DNA. J Biol Chem. 1991 Aug 15;266(23):15160–15169. [PubMed] [Google Scholar]
- Doerfler W. Abortive infection and malignant transformation by adenoviruses: integration of viral DNA and control of viral gene expression by specific patterns of DNA methylation. Adv Virus Res. 1991;39:89–128. doi: 10.1016/s0065-3527(08)60793-9. [DOI] [PubMed] [Google Scholar]
- Doerfler W. DNA methylation and gene activity. Annu Rev Biochem. 1983;52:93–124. doi: 10.1146/annurev.bi.52.070183.000521. [DOI] [PubMed] [Google Scholar]
- Doerfler W. Patterns of DNA methylation--evolutionary vestiges of foreign DNA inactivation as a host defense mechanism. A proposal. Biol Chem Hoppe Seyler. 1991 Aug;372(8):557–564. [PubMed] [Google Scholar]
- Doerfler W. Patterns of de novo DNA methylation and promoter inhibition: studies on the adenovirus and the human genomes. EXS. 1993;64:262–299. doi: 10.1007/978-3-0348-9118-9_12. [DOI] [PubMed] [Google Scholar]
- Drake J. W., Allen E. F., Forsberg S. A., Preparata R. M., Greening E. O. Genetic control of mutation rates in bacteriophageT4. Nature. 1969 Mar 22;221(5186):1128–1132. [PubMed] [Google Scholar]
- Fearon E. R., Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990 Jun 1;61(5):759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- Flavell A. J., Brierley C. The termini of extrachromosomal linear copia elements. Nucleic Acids Res. 1986 May 12;14(9):3659–3669. doi: 10.1093/nar/14.9.3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartler S. M., Riggs A. D. Mammalian X-chromosome inactivation. Annu Rev Genet. 1983;17:155–190. doi: 10.1146/annurev.ge.17.120183.001103. [DOI] [PubMed] [Google Scholar]
- Gautsch J. W., Wilson M. C. Delayed de novo methylation in teratocarcinoma suggests additional tissue-specific mechanisms for controlling gene expression. Nature. 1983 Jan 6;301(5895):32–37. doi: 10.1038/301032a0. [DOI] [PubMed] [Google Scholar]
- Holliday R. The inheritance of epigenetic defects. Science. 1987 Oct 9;238(4824):163–170. doi: 10.1126/science.3310230. [DOI] [PubMed] [Google Scholar]
- Honess R. W., Gompels U. A., Barrell B. G., Craxton M., Cameron K. R., Staden R., Chang Y. N., Hayward G. S. Deviations from expected frequencies of CpG dinucleotides in herpesvirus DNAs may be diagnostic of differences in the states of their latent genomes. J Gen Virol. 1989 Apr;70(Pt 4):837–855. doi: 10.1099/0022-1317-70-4-837. [DOI] [PubMed] [Google Scholar]
- Hu L. F., Minarovits J., Cao S. L., Contreras-Salazar B., Rymo L., Falk K., Klein G., Ernberg I. Variable expression of latent membrane protein in nasopharyngeal carcinoma can be related to methylation status of the Epstein-Barr virus BNLF-1 5'-flanking region. J Virol. 1991 Mar;65(3):1558–1567. doi: 10.1128/jvi.65.3.1558-1567.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JOSSE J., KAISER A. D., KORNBERG A. Enzymatic synthesis of deoxyribonucleic acid. VIII. Frequencies of nearest neighbor base sequences in deoxyribonucleic acid. J Biol Chem. 1961 Mar;236:864–875. [PubMed] [Google Scholar]
- Jansson A., Masucci M., Rymo L. Methylation of discrete sites within the enhancer region regulates the activity of the Epstein-Barr virus BamHI W promoter in Burkitt lymphoma lines. J Virol. 1992 Jan;66(1):62–69. doi: 10.1128/jvi.66.1.62-69.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P. A., Rideout W. M., 3rd, Shen J. C., Spruck C. H., Tsai Y. C. Methylation, mutation and cancer. Bioessays. 1992 Jan;14(1):33–36. doi: 10.1002/bies.950140107. [DOI] [PubMed] [Google Scholar]
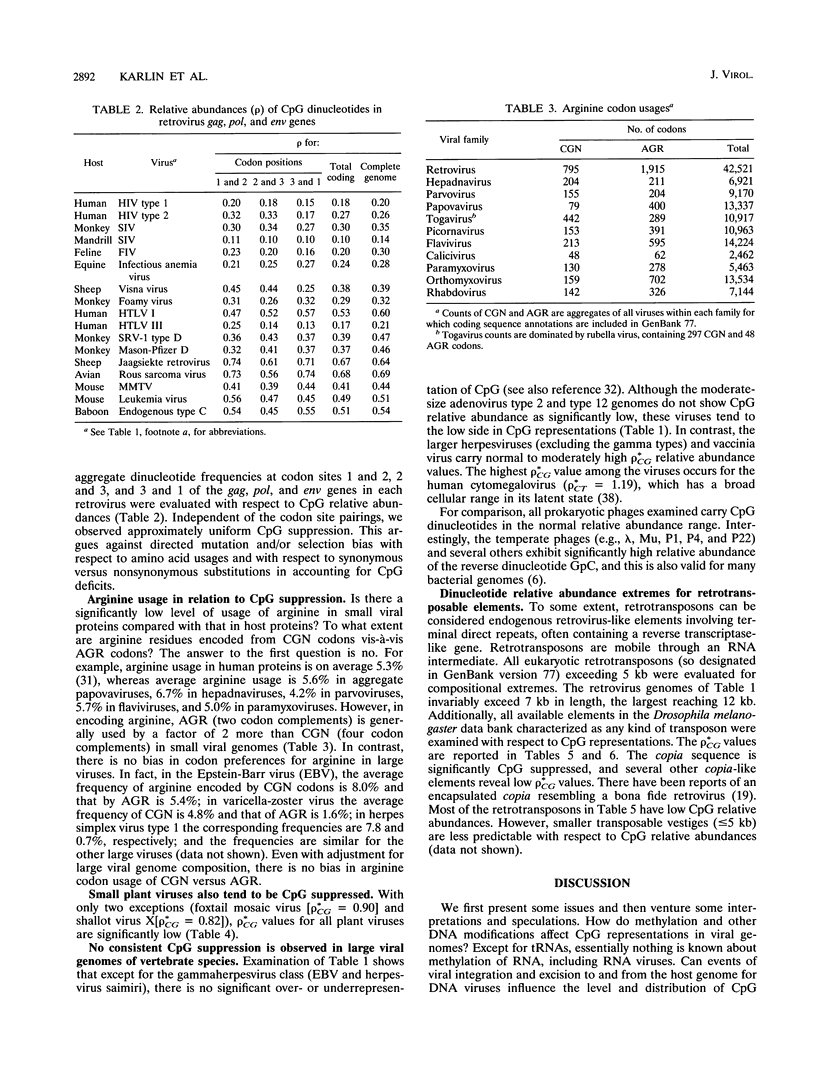
- Karlin S., Blaisdell B. E., Bucher P. Quantile distributions of amino acid usage in protein classes. Protein Eng. 1992 Dec;5(8):729–738. doi: 10.1093/protein/5.8.729. [DOI] [PubMed] [Google Scholar]
- Karlin S., Mocarski E. S., Schachtel G. A. Molecular evolution of herpesviruses: genomic and protein sequence comparisons. J Virol. 1994 Mar;68(3):1886–1902. doi: 10.1128/jvi.68.3.1886-1902.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshet I., Lieman-Hurwitz J., Cedar H. DNA methylation affects the formation of active chromatin. Cell. 1986 Feb 28;44(4):535–543. doi: 10.1016/0092-8674(86)90263-1. [DOI] [PubMed] [Google Scholar]
- Kricker M. C., Drake J. W., Radman M. Duplication-targeted DNA methylation and mutagenesis in the evolution of eukaryotic chromosomes. Proc Natl Acad Sci U S A. 1992 Feb 1;89(3):1075–1079. doi: 10.1073/pnas.89.3.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchini A., Tomkinson B., Cohen J. I., Kieff E. BHRF1, the Epstein-Barr virus gene with homology to Bc12, is dispensable for B-lymphocyte transformation and virus replication. J Virol. 1991 Nov;65(11):5991–6000. doi: 10.1128/jvi.65.11.5991-6000.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minarovits J., Minarovits-Kormuta S., Ehlin-Henriksson B., Falk K., Klein G., Ernberg I. Host cell phenotype-dependent methylation patterns of Epstein-Barr virus DNA. J Gen Virol. 1991 Jul;72(Pt 7):1591–1599. doi: 10.1099/0022-1317-72-7-1591. [DOI] [PubMed] [Google Scholar]
- Morrison J. M., Keir H. M., Subak-Sharpe H., Crawford L. V. Nearest neighbour base sequence analysis of the deoxyribonucleic acids of a further three mammalian viruses: Simian virus 40, human papilloma virus and adenovirus type 2. J Gen Virol. 1967 Jan;1(1):101–108. doi: 10.1099/0022-1317-1-1-101. [DOI] [PubMed] [Google Scholar]
- Niwa O., Yokota Y., Ishida H., Sugahara T. Independent mechanisms involved in suppression of the Moloney leukemia virus genome during differentiation of murine teratocarcinoma cells. Cell. 1983 Apr;32(4):1105–1113. doi: 10.1016/0092-8674(83)90294-5. [DOI] [PubMed] [Google Scholar]
- Peck L. J., Nordheim A., Rich A., Wang J. C. Flipping of cloned d(pCpG)n.d(pCpG)n DNA sequences from right- to left-handed helical structure by salt, Co(III), or negative supercoiling. Proc Natl Acad Sci U S A. 1982 Aug;79(15):4560–4564. doi: 10.1073/pnas.79.15.4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rideout W. M., 3rd, Coetzee G. A., Olumi A. F., Jones P. A. 5-Methylcytosine as an endogenous mutagen in the human LDL receptor and p53 genes. Science. 1990 Sep 14;249(4974):1288–1290. doi: 10.1126/science.1697983. [DOI] [PubMed] [Google Scholar]
- Schetter C., Grünemann B., Hölker I., Doerfler W. Patterns of frog virus 3 DNA methylation and DNA methyltransferase activity in nuclei of infected cells. J Virol. 1993 Dec;67(12):6973–6978. doi: 10.1128/jvi.67.12.6973-6978.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selker E. U. DNA methylation and chromatin structure: a view from below. Trends Biochem Sci. 1990 Mar;15(3):103–107. doi: 10.1016/0968-0004(90)90193-f. [DOI] [PubMed] [Google Scholar]
- Selker E. U. Premeiotic instability of repeated sequences in Neurospora crassa. Annu Rev Genet. 1990;24:579–613. doi: 10.1146/annurev.ge.24.120190.003051. [DOI] [PubMed] [Google Scholar]
- Shpaer E. G., Mullins J. I. Selection against CpG dinucleotides in lentiviral genes: a possible role of methylation in regulation of viral expression. Nucleic Acids Res. 1990 Oct 11;18(19):5793–5797. doi: 10.1093/nar/18.19.5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson R. T. Nucleosome positioning: occurrence, mechanisms, and functional consequences. Prog Nucleic Acid Res Mol Biol. 1991;40:143–184. doi: 10.1016/s0079-6603(08)60841-7. [DOI] [PubMed] [Google Scholar]
- Smith D. B., Inglis S. C. The mutation rate and variability of eukaryotic viruses: an analytical review. J Gen Virol. 1987 Nov;68(Pt 11):2729–2740. doi: 10.1099/0022-1317-68-11-2729. [DOI] [PubMed] [Google Scholar]
- Subak-Sharpe H., Bürk R. R., Crawford L. V., Morrison J. M., Hay J., Keir H. M. An approach to evolutionary relationships of mammalian DNA viruses through analysis of the pattern of nearest neighbor base sequences. Cold Spring Harb Symp Quant Biol. 1966;31:737–748. doi: 10.1101/sqb.1966.031.01.094. [DOI] [PubMed] [Google Scholar]
- Sutter D., Westphal M., Doerfler W. Patterns of integration of viral DNA sequences in the genomes of adenovirus type 12-transformed hamster cells. Cell. 1978 Jul;14(3):569–585. doi: 10.1016/0092-8674(78)90243-x. [DOI] [PubMed] [Google Scholar]
- Tazi J., Bird A. Alternative chromatin structure at CpG islands. Cell. 1990 Mar 23;60(6):909–920. doi: 10.1016/0092-8674(90)90339-g. [DOI] [PubMed] [Google Scholar]
- Tilghman S. M. DNA methylation: a phoenix rises. Proc Natl Acad Sci U S A. 1993 Oct 1;90(19):8761–8762. doi: 10.1073/pnas.90.19.8761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers A. A., Schwabe J. W. Spurring on transcription? Curr Biol. 1993 Dec 1;3(12):898–900. doi: 10.1016/0960-9822(93)90231-c. [DOI] [PubMed] [Google Scholar]
- Willis D. B., Granoff A. Frog virus 3 DNA is heavily methylated at CpG sequences. Virology. 1980 Nov;107(1):250–257. doi: 10.1016/0042-6822(80)90290-1. [DOI] [PubMed] [Google Scholar]
- Youssoufian H., Hammer S. M., Hirsch M. S., Mulder C. Methylation of the viral genome in an in vitro model of herpes simplex virus latency. Proc Natl Acad Sci U S A. 1982 Apr;79(7):2207–2210. doi: 10.1073/pnas.79.7.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]



