Abstract
Mainly based on various inhibitor studies previously performed, amidases came to be regarded as sulfhydryl enzymes. Not completely satisfied with this generally accepted interpretation, we performed a series of site-directed mutagenesis studies on one particular amidase of Rhodococcus rhodochrous J1 that was involved in its nitrile metabolism. For these experiments, the recombinant amidase was produced as the inclusion body in Escherichia coli to greatly facilitate its recovery and subsequent purification. With regard to the presumptive active site residue Cys203, a Cys203 → Ala mutant enzyme still retained 11.5% of the original specific activity. In sharp contrast, substitutions in certain other positions in the neighborhood of Cys203 had a far more dramatic effect on the amidase. Glutamic acid substitution of Asp191 reduced the specific activity of the mutant enzyme to 1.33% of the wild-type activity. Furthermore, Asp191 → Asn substitution as well as Ser195 → Ala substitution completely abolished the specific activity. It would thus appear that, among various conserved residues residing within the so-called signature sequence common to all amidases, the real active site residues are Asp191 and Ser195 rather than Cys203. Inasmuch as an amide bond (CO-NH2) in the amide substrate is not too far structurally removed from a peptide bond (CO-NH-), the signature sequences of various amidases were compared with the active site sequences of various types of proteases. It was found that aspartic acid and serine residues corresponding to Asp191 and Ser195 of the Rhodococcus amidase are present within the active site sequences of aspartic proteinases, thus suggesting the evolutionary relationship between the two.
Keywords: hydrolase, aspartic acid, nitrile, serine, Rhodococcus
An amide bond is of considerable importance in biochemistry because many C-terminal amide-containing peptides act as hormones (1). Amidohydrolases, amide bond-cleaving enzymes, exist in both prokaryotic and eukaryotic forms. According to the categorical numbering system of EC (2) that uses such properties as substrate specificity and physicochemical characteristics as criteria, amidohydrolases have been divided into two major types: 77 were included in the EC 3.5.1 category (EC 3.5.1.1–3.5.1.77), and 14 were placed under EC 3.5.2 (EC 3.5.2.1–3.5.2.14). Among them, one amidase (EC 3.5.1.4) in particular (with which we are here concerned) recently has received much attention in diverse fields such as neurobiochemistry, plant physiology, and applied microbiology. This enzyme, which catalyzes hydrolysis of an amide to an acid and ammonium, has been shown to be involved in the following areas: (i) processes inactivating fatty acid amides as neuronally active signaling molecules (3); (ii) biosynthesis of indoleacetic acid as an important plant hormone (4, 5); and (iii) enzymatic production in an industrial scale of marketable compounds (6–8). A series of studies on the biosynthesis by this amidase of indoleacetic acid finally led to the fruitful development of the transformation system that converts callus tissues to individual plants (9), and studies on the microbial metabolism of carbon–nitrogen-containing compounds by amidase and nitrile hydratase led to the industrial scale enzymatic production of acrylamide and nicotinamide (6, 7).
Our amidase (10) is involved in nitrile metabolism of an industrially adapted Rhodococcus rhodochrous J1 strain (6, 7), and, in this organism, the amidase structural gene exists in close linkage with two other structural genes encoding cobalt transporter (11) and cobalt-containing nitrile hydratase (12). This Rhodococcus amidase (10) shares sequence similarities with indoleacetamide hydrolase (indoleacetic acid-synthesizing enzyme) (4, 5), oleamide hydrolase (3), a vitamin D3 hydroxylase-associated protein (13, 14), a nylon–oligomer-degrading EI enzyme (6-aminohexanoate-cyclic dimer hydrolase; EC 3.5.2.12) (15, 16), and other amidases that are coupled with nitrile hydratases to be involved in microbial nitrile metabolism (17–21). These similarities are particularly strong within their common signature sequences (3, 13, 17). So far, members of the so-called amidase family generally have been classified as belonging to a branch of the sulfhydryl enzymes based on the results of the experiments using inhibitors (8, 18). Nevertheless, active amino acid residues have not been identified in any of the enzymes belonging to the amidase family. Identification of the active site will certainly help in elucidating its unique specificity for each amide substrate as well as its reaction mechanism and subsequently help us to understand its physiological functions. Therefore, we have attempted to identify active amino acid residues in the R. rhodochrous J1 amidase.
On the other hand, the aspartic proteinase that is one of the peptide bond-cleaving enzymes is used as a therapeutic target of AIDS (22) and in cheese manufacture (23). Nevertheless, no one has reported the evolutionary relationship between amide bond-cleaving enzymes (including amidase) and peptide bond-cleaving enzymes (including aspartic proteinase), despite their physiological importance. We also describe here a surprising evolutionary relationship between the amidase family and the aspartic proteinase family.
MATERIALS AND METHODS
Strains, Plasmids, and DNA Manipulation.
Escherichia coli JM109 (24) was used as the host strain for recombinant plasmids and for phage M13 mp18/19 propagation (24). The plasmid pALJ30 carrying the amidase gene of R. rhodochrous J1 in the 1.6-kb SacI–HindIII fragment on pUC18 (10) was used for expression and site-directed mutagenesis of the amidase gene. DNA manipulation was performed essentially as described by Sambrook et al. (24). The DNA sequence was determined by the dideoxynucleotide chain termination method (25).
Site-Directed Mutagenesis.
Four oligonucleotides were synthesized for mutagenesis by the phosphorothioate-based strategy of Taylor et al. (26): 5′-GTCGGCGGCAACCAGGG-3′ for replacement of Asp191 by Asn, 5′-GGCGGCGAGCAGGGCG-3′ for replacement of Asp191 by Glu, 5′-GGGCGGTGCGATCCG-3′ for replacement of Ser195 by Ala, and 5′-CCGCGTTCGCCGGCATCGTC-3′ for replacement of Cys203 by Ala; underlining indicates mismatched bases. To prepare single-stranded DNA, the SacI and HindIII sites of replicative form M13 mp19DNA were ligated with the 1.6-kb SacI–HindIII fragment containing the Rhodococcus amidase gene isolated from pALJ30, which was constructed for the production of amidase in E. coli JM109 (10). The desired mutants were sequenced to identify each mutation and confirm their fidelity.
Preparation of Cell Extracts and Enzyme Assays.
The E. coli transformant carrying pALJ30 with the R. rhodochrous J1 wild-type amidase gene (10) was cultured at 28°C for 8 h in 100 ml of 2 × YT medium (1.6% Tryptone/1% yeast extract/0.5% NaCl) containing ampicillin in a 500-ml shaking flask with isopropyl-β-d-thiogalactopyranoside added to a final concentration of 1 mM to induce the lac promoter. The cells were harvested by centrifugation, suspended in 3 ml of 0.1 M potassium phosphate buffer (pH 7.5) containing 1 mM DTT, and disrupted by sonication for 5 min (Insonator model 201M, Kubota, Japan) to prepare the cell-free extracts. The extracts of the transformants carrying D191N, D191E, S195A ,or C203A mutant amidase also were prepared under the same conditions as for the wild-type amidase. The extracts were then centrifuged at 12,000 × g for 30 min, and the amidase activities of each supernatant were measured by the same method as described (10). One unit of amidase activity was defined as the amount of enzyme that catalyzed the formation of 1 μmol of benzoic acid/min (from benzamide as a substrate) under the above conditions. The specific activity was expressed as units per milligram of protein.
Purification of Wild-Type and Mutant Amidases from E. coli Transformants.
The E. coli transformant carrying pALJ30 (10) was cultured at 37°C for 18 h in 2× YT medium containing ampicillin and isopropyl-β-d-thiogalactopyranoside, harvested by centrifugation, and disrupted by sonication. The extracts were then centrifuged at 12,000 × g for 30 min, and the resultant precipitates were solubilized with the lysis buffer (50 mM Tris/100 mM NaCl/1 mM EDTA/0.5% Triton X-100) to remove the membrane fraction in a soluble form by centrifugation at 17,000 × g for 15 min. The resultant inclusion bodies as precipitates were solubilized by 8 M urea in the dialysis buffer (30 mM Tris/30 mM NaCl/1 mM DTT/glycerol, pH 7.5) and dialyzed against the dialysis buffer containing 4 M urea for 12 h and then against only the dialysis buffer for 24 h at 4°C. The renatured wild-type amidase was easily purified from the dialyzed solution by the following two-step procedures at 0–4°C using potassium phosphate buffer (pH 7.5) containing 1 mM DTT and 20% (mass/vol) glycerol unless otherwise specified. The dialyzed enzyme solution was applied to a Mono-Q HR 5/5 column equilibrated with 10 mM buffer, which was attached to a fast protein liquid chromatography system (Pharmacia). After the column was washed with the same buffer, the enzyme was eluted by increasing the ionic strength of KCl in a linear fashion from 0.1 to 0.5 M in the same buffer at a flow rate of 0.5 ml/min. The active fractions were pooled. Sodium ammonium sulfate was added to the resulting enzyme solution to give 45% (mass/vol) saturation. After stirring for 4 h or more, the precipitate was removed by centrifugation, and ammonium sulfate was added to the supernatant to give 60% (mass/vol) saturation. The suspension was then centrifuged, and the pellet was dissolved in 0.1 M buffer, followed by dialysis for 24 h against 10 mM buffer.
Each D191N, D191E, S195A, and C203A mutant amidase also was purified from each mutant amidase-producing transformant according to the same procedure used for the purification of the wild-type amidase described above.
RESULTS AND DISCUSSION
Site-Directed Mutagenesis and Enzyme Assay in the Supernatants of the E. coli Transformant Extracts.
The Cys203 residue in the Rhodococcus amidase (10) was at first selected for the mutagenesis experiment because, thus far, the amidase had generally been classified as one of the sulfhydryl enzymes (8, 18), and this cysteine residue is conserved in most of the amidase family (3, 13, 17) whereas other three cysteine residues (Cys141, Cys145, and Cys464) of the R. rhodochrous J1 amidase are not conserved in the corresponding sequences of most of the family. The Asp191 and Ser195 residues were next selected because they are absolutely conserved in the common signature sequence of the amidase family (3, 13, 17). By site-directed mutagenesis, we constructed four mutant amidases, in which Asp191 was replaced by Asn as well as Glu while Ser195 and Cys203 were replaced by Ala. The identity of each mutant was confirmed by determining the complete nucleotide sequence of the mutant gene. Each mutant enzyme was named using the single letter code for amino acids to indicate the substitutions made. The E. coli transformant cells containing each mutant amidase gene were cultured at 28°C using the expression system that we established previously for the wild-type amidase (10). Amidase activities in the supernatants of cell-free extracts prepared from the cells were measured.
The C203A mutant enzyme had specific activity of 0.0306 units/mg in the supernatant of the cell-free extracts, which was 10.8% of the activity of the wild-type amidase (0.283 units/mg: 100%) (10). On the other hand, the D191E mutant enzyme had specific activity of 0.0021 units/mg (0.75% of the wild-type amidase activity) in the supernatant. Each mutant enzyme containing the D191N substitution and the S195A substitution did not exhibit any amidase activity at all.
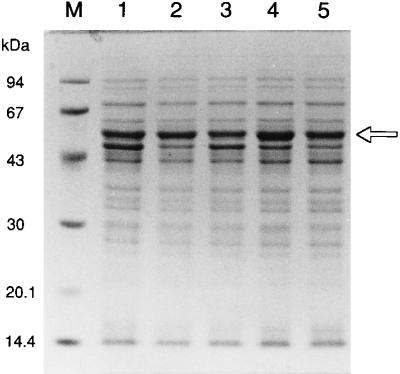
The corresponding amidase bands were detected in the supernatants of the cell-free extracts containing each mutant enzyme in SDS/PAGE (Fig. 1); each positive band in Western blot analysis with anti-amidase antiserum (12) (data not shown) also was found to be the Rhodococcus amidase by determination of the N-terminal 25-amino acid sequences on a gas-phase amino acid sequencer (Applied Biosystems, model 470A).
Figure 1.
SDS/PAGE of the R. rhodochrous J1 wild-type and mutant amidases in the supernatants of extracts of each E. coli transformant. The supernatants of extracts prepared from each transformant by the procedure described in Materials and Methods were loaded onto SDS/PAGE gels. The amidase bands are indicated by an arrow. Lanes: M, molecular mass standards; 1, the wild-type amidase; 2, the D191N amidase; 3, the D191E amidase; 4, the S195A amidase; and 5, the C203A amidase.
Denaturation–Renaturation of Inclusion Bodies of Wild-Type and Mutant Amidases and Their Purification.
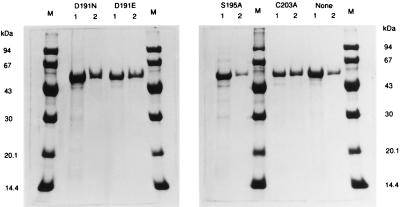
It was believed that findings on the cell-free extract supernatants described above had to be verified on purified amidases, the wild-type as well as mutants. Therefore, we attempted to purify these mutant amidases. However, the expression system in E. coli used above was not suitable for amidase overexpression. Although it produced the wild-type and mutant amidases in soluble active form, the amount produced was not very great, resulting in the long purification procedure as reported (10). Fortunately, it was found that the enzyme became hyperexpressed to form inclusion bodies, albeit in the inactive form, when the E. coli transformant containing pALJ30 (10) was cultured at 37°C for 18 h. The amount of the amidase corresponded to >90% of the total insoluble proteins (Fig. 2, lane 1). The inclusion bodies were solubilized by urea, followed by their renaturation through dialysis as described in Materials and Methods. At the end, the wild-type amidase with a specific activity (11.6 units/mg) nearly equal to that (12.2 units/mg) of the R. rhodochrous J1 amidase purified from a soluble fraction (10) was obtained. The enzyme was easily purified (Fig. 2, lane 2) from the dialyzed enzyme solution through a two-step procedure: column chromatography and ammonium sulfate fractionation as described in Materials and Methods.
Figure 2.
SDS/PAGE gel showing the purification stages of the wild-type and mutant amidases from the insoluble fraction of extracts in the E. coli transformant. Lanes D191N, D191E, S195A, C203A, and None indicate the D191N, D191E, S195A, and C203A mutant amidases and the wild-type amidase, respectively. Lanes M were loaded with molecular mass standards. Lanes 1, inclusion bodies; lanes 2, purified enzymes.
We next overexpressed and purified each D191N, D191E, S195A, and C203A mutant amidase under the same conditions as in the wild-type amidase (Fig. 2). Their circular dichroism spectra in the far UV region and molecular mass were almost identical to those of the wild-type amidase (data not shown), demonstrating that essentially no major change in the overall conformation occurred in these mutant enzymes. Specific activities of both purified D191N and S195A mutant enzymes were less than the detection threshold, even when large amounts of the enzymes were used in the reaction for 15 h. On the other hand, the purified D191E mutant enzyme and the purified C203A mutant enzyme had specific activities of 0.162 and 1.40 units/mg, respectively, which were 1.33% and 11.5%, respectively, of the activity shown by the purified wild-type amidase [12.2 units/mg (10): 100%]. For benzamide as a substrate, the Km value of the purified D191E mutant enzyme was 0.68 mM; that of the purified C203A enzyme was 0.16 mM, which was similar to that of the purified wild-type amidase [0.15 mM (10)].
These findings indicated that Asp191 and Ser195 would be crucial to the catalytic activity whereas Cys203 was not. However, Cys203 was apparently involved in enhancement of the catalytic activity. The above is in agreement with the finding that Cys203 is not completely conserved by every member of the amidase family (10) (Fig. 3) and with the finding that the purified wild-type amidase shows a strong resistance toward thiol reagents (10). The suggestion that amidases are the sulfhydryl enzyme (8, 18) also has been made based upon the inhibitor studies performed upon the EI enzyme of Flavobacterium (15). The sensitivities of other amidases to thiol reagents might be attributable to the close proximity of the active aspartic acid and serine residues to cysteine; the access of thiol reagents to the cysteine residue of interest may cause steric hindrance to the function of the former active two residues. As to Ser195 of the Rhodococcus amidase, this amidase was not inactivated by either diisopropylfluorophosphate or phenylmethanesulfonyl fluoride, which were affinity labels for serine proteases (27). Although this finding seems to be in disagreement with the crucial role of Ser195 shown by the site-directed mutagenesis, such inconsistency is known in several enzymes, e.g., leader peptidase, which removes signal peptides from exported proteins, is a serine protease despite insensibility to diisopropylfluorophosphate or phenylmethanesulfonyl fluoride (28). Thus, it appears that the active site serine of amidases plays a role different from that played by the active site serine of serine proteases.
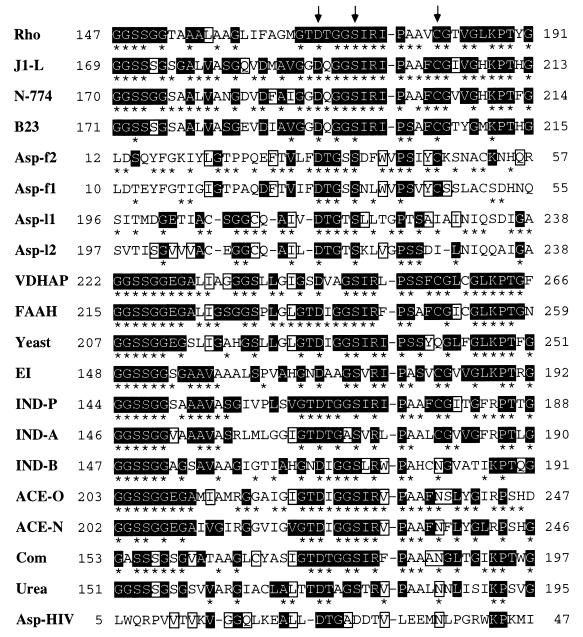
Figure 3.
Alignment over the amidase signature sequence region (3, 13, 17) of all members of the amidase family previously reported with the aspartic proteinases. From top to bottom (numbers of the total amino acids of each overall sequence and their accession numbers of databases are given in brackets just after each enzyme name): Rho, amidase from Rhodococcus sp. [462 residues, GenBank M74531 (17)]; J1-L, amidase from R. rhodochrous J1 [515 residues, GenBank D16207 (10)]; N-774, amidase from Rhodococcus sp. N-774 (whose sequence is identical with that of the Brevibacterium R312 amidase) [521 residues, GenBank X54074 or M60264 (19, 20)]; B23, amidase from Pseudomonas chlororaphis B23 [506 residues, GenBank D90216 (21)]; Asp-f2, the N-terminal region of calf chymosin [323 residues, PIR CMBO (43, 44)]; Asp-f1, the N-terminal region of porcine pepsin [325 residues, GenBank J04601 (45)]; Asp-l1, the C-terminal region of porcine pepsin [325 residues, GenBank J04601 (45)]; Asp-l2, the C-terminal region of calf chymosin [323 residues, PIR CMBO (43, 44)]; VDHAP, a vitamin D3 hydroxylase-associated protein from cockerel [464 residues, GenBank U00694 (13)]; FAAH, oleamide hydrolase from rat [579 residues, GenBank U72497 (3)]; Yeast, putative amidase from Saccharomyces cerevisiae [549 residues, GenBank X56043 (46) (only this putative amidase is described here as a representative of putative amidases; two other putative sequences of Caenorhabditis elegans are not shown here)]; EI, EI enzyme from Flavobacterium [493 residues, GenBank M26953 (15)]; IND-P, indoleacetamide hydrolase from P. savastanoi [455 residues, GenBank M11035 (4)]; IND-A, indoleacetamide hydrolase from Agrobacterium tumefaciens [467 residues, PIR Q2AGAT (5)]; IND-B, indoleacetamide hydrolase from Bradyrhizobium japonicum [465 residues, GenBank X15117 (47)]; ACE-O, acetamidase from Aspergillus oryzae [545 residues, GenBank D10492 (48)]; ACE-N, acetamidase from Aspergillus nidulans [548 residues, GenBank M16371 (49)]; Com, amidase from Comamonas acidovorans KPO-2771–4 [473 residues (8)]; Urea, urea amidolyase from Candida utilis [1830 residues, PRF 2009329A (34)]; Asp–HIV, aspartic proteinase from HIV type-1 [102 residues, GenBank HIVU53613 (50)]. Arrows indicate the amino acid residues that correspond to Asp191, Ser195, and Cys203 of the R. rhodochrous J1 amidase examined in this experiment. Numbers on the left and right indicate the position of the amino acid residue in each molecule. Gaps (-) were inserted to increase sequence similarity. Identical residues are denoted by asterisks between neighboring sequences. Residues highlighted in reverse type are conserved across both amidases and aspartic proteinases in at least four of 20 sequences, and the first four residues GGSS highlighted in reverse type in most of the amidases do not appear in the aspartic proteinases with the exception of Ser14 in Asp-f2. Except the residues highlighted in reverse type, identical amino acid residues, all of which are not conserved between both aspartic proteinase and amidase families, are boxed.
Sequence Similarity Between Amidase and Aspartic Proteinase Around Each Active Aspartic Acid Residue.
Inasmuch as we have identified Asp191 and Ser195 located within the signature sequence of the amidase family as the active site residues, we made the homology search using the available database and found the significant similarity between the signature sequence of the amidase family and either of the two active site sequences of aspartic proteinases such as pepsin and chymosin (29) (Fig. 3). Aspartic proteinases have two active aspartic acid residues; one in the amino-terminal half (position 32 in the porcine pepsin-numbering system used in Fig. 3) and the other in the carboxyl-terminal half (position 215). The presence of two active sites was thought to be a consequence of tandem duplication (30, 31). HIV aspartic proteinase is a little bit different from the above-noted aspartic proteinases in that it is endowed with only one active site. Yet, it functions as a homodimer, leading to the conformation of two active sites (22). When the region around the active site residue Asp191 of the R. rhodochrous J1 amidase was compared with that of the aspartic proteinases, there was a gap in the region (residue positions 198–199) of the R. rhodochrous J1 amidase and in the region (positions 32–33) of the HIV protease in contrast with nongap in the corresponding region of porcine pepsin and calf chymosin. Furthermore, the position corresponding to the active site Ser195 was occupied by another aspartic acid in the HIV protease (Fig. 3).
The region in the amino-terminal half of indoleacetamide hydrolases (e.g., positions 62–224 of the Pseudomonas savastanoi hydrolase) (4, 5) showed some similarity to the signature sequence of certain amidases that are functionally coupled with nitrile hydratases (shown as Rho, J1-L, N-774, and B23 in Fig. 3). In fact, within the region representing positions 62–224 of the P. savastanoi hydrolase (4), we seemed to have identified active site aspartic acid corresponding to Asp191 of the Rhodococcus amidase. The carboxyl-terminal half of indoleacetamide hydrolases (e.g., positions 225–455 of the Pseudomonas hydrolase), on the other hand, showed almost no similarity to any part of amidases (10).
The location of the active aspartic acid in the overall amino acid sequence of the R. rhodochrous J1 amidase also corresponded well to the second active aspartic acid of the aspartic proteinases and that of the possible active aspartic acid in the other amide-degrading enzymes (Fig. 3). The recognizable homology between sequences surrounding the active aspartic acid among the amide-degrading enzymes as well as the proteases suggested that such homology is of functional and evolutionary significance. Both amidase and aspartic proteinase cleave a carbon–nitrogen bond of a substrate, despite a difference in adjacent structures of each scissile bond; the former acts on amide bonds whereas the latter acts on peptide bonds. Because the reaction formula of amidase is somewhat similar to that of aspartic proteinase and the R. rhodochrous J1 amidase is a homodimer enzyme like the HIV protease, there is a possibility that two catalytic aspartic acid residues of the former enzyme, each of which comes from a different subunit, form a catalytic site, as in the latter enzyme. However, the catalytic mechanism of amidases would not be analogous to the known mechanism of aspartic proteinases because the R. rhodochrous J1 amidase was not inactivated by N-diazoacetyl-l-phenylalanine methyl ester, 1,2-epoxy-3-(p-nitrophenoxy)propane, and pepstatin (data not shown), which were known to be inhibitors of the aspartic proteinases. Amidases would operate by an acyl–enzyme mechanism (3, 32) when showing acyl transferase activity (10, 32) whereas the reaction by aspartic proteinases is thought to proceed via a tetrahedral intermediate (different from an acyl–enzyme intermediate) (30), rather than an oxyanion derivative of the peptide as in serine protease catalysis (27). We speculate that Asp191 of the Rhodococcus amidase may function as a general base abstracting the proton from the Ser195 hydroxyl sidechain, allowing the serine to act as a nucleophile for the carbonyl group of the amide bond within the substrate. Further studies are required to clarify the catalytic mechanism of the amidase because we cannot rule out a possibility that minor structural changes generated by the amino acid substitutions resulted in the complete loss of the enzyme activity.
The amidase family (3, 10, 13, 17) shows a significant sequence similarity with the EI enzyme (15) that is involved in nylon–oligomer degradation, in combination with the EII enzyme, in Flavobacterium (16). Tsuchiya et al. (15) reported that the EI enzyme has shown a sequence similarity with the EII enzyme (6-aminohexanoate-dimer hydrolase; EC 3.5.1.46) particularly in the short region [around the active serine residue in the latter (33)], which locally corresponded to that containing the active Ser195 in the R. rhodochrous J1 amidase. They also suggested that the EI enzyme might be a serine enzyme and the formation of a superfamily consisting of this enzyme and active site–serine DD peptidases (class C β-lactamases) that catalyzes the attack of a C-terminal d-alanyl-d-alanine peptide bond in peptidoglycan precursors involved in bacterial cell wall metabolism. The amidase family also shows similarity with the putative allophanate hydrolase (EC 3.5.1.54) [which is a part of a multifunctional enzyme, urea amidolyase (34)] from Candida utilis. The total number of the urea amidolyase sequence (1830 amino acid residues), however, was far greater than that of the amidases, aspartic proteinases, and others (323–579 residues) listed in Fig. 3. The HIV protease, on the other hand, was comprised of only 102 residues.
Proteases are mainly divided into four categories: cysteine proteases, serine proteases, aspartic proteinases, and metal proteases. However, much still remains to be done to clarify evolutionary relationships among them. We could not observe sequence similarities between amidase and proteases other than the aspartic proteinases. It is interesting that the amidase reaction would proceed through an acyl–enzyme intermediate in contrast with that of aspartic proteinase despite the significant similarity within the active site sequences between the two enzymes while there is a possibility that the common amino acid residues have been selected from random sequences by functional constraint through the evolution, in contrast with a possibility that the two sequences have been diverged from the common antecedent. Further studies on the amidase family from standpoints of its reaction mechanism and its three-dimensional structure could provide information about their evolutionary relationships.
The expression of the R. rhodochrous J1 amidase gene is regulated by nhlC and nhlD [positive and negative regulators, respectively (12)]; of the two, nhlC shows a sequence similarity with a regulator amiC for the expression of an aliphatic amidase (in Pseudomonas aeruginosa) (35). This aliphatic amidase is classified as one group belonging to the C–N hydrolase superfamily (36, 37). This superfamily included nitrilases that convert the cyano group of a nitrile into a carboxylic acid (38, 39) and cyanide hydratases that convert a cyano group of a cyanide into an amide (40, 41). Members of this superfamily shared several signature sequences of their own, one of which contained an invariant cysteine identified by us as an active amino acid residue (38, 42). However, there were no homologies between the amidase family that included the Rhodococcus amidase and the C–N hydrolase superfamily that included the Pseudomonas aliphatic amidase. Moreover, we have never found sequence similarity among the amidase family and all the amidohydrolases (whose sequences were reported) other than the amidases listed in Fig. 3. Inasmuch proteases have been shown to be comprised of four major groups on the basis of having different active sites, it would not be a surprise if, in the future, amidohydrolases too are shown to be comprised of several distinct groups, e.g., cysteine amidohydrolase, serine amidohydrolase, aspartic amidohydrolase, and metal amidohydrolase. The amidase described in this paper can be classified either as a serine amidohydrolase or as an aspartic amidohydrolase whereas the Pseudomonas aliphatic amidase already noted becomes a cysteine amidohydrolase.
The gene encoding indoleacetamide hydrolase (4, 5) is coupled with tryptophan 2-monooxygenase in the biosynthesis of indoleacetic acid from tryptophan in phytopathogenic bacteria. This indoleacetamide hydrolase is expressed in plants that are infected with these bacteria, the production of indoleacetic acid triggering continuous proliferation of the transformed cells. It also should be noted that amidases such as oleamide hydrolase are encoded in the mammalian genome. Studies on the amidase as well as aspartic proteinase families are relevant in understanding not only the distribution of both families in viruses, microorganisms, plants, and mammals but also the nature of their evolutionary relationships.
Acknowledgments
We thank Dr. S. Ohno (Beckman Research Institute of the City of Hope) for his helpful suggestions and critical reading of this manuscript. This work was supported in part by a Grant-in-Aid for Scientific Research from the Ministry of Education, Science, and Culture of Japan and by a grant from The Naito Foundation.
References
- 1.Bradbury A F, Smyth D G. Trends Biochem Sci. 1991;16:112–115. doi: 10.1016/0968-0004(91)90044-v. [DOI] [PubMed] [Google Scholar]
- 2.Schomburg D, Salzmann M, editors. Enzyme Handbook 4. Berlin: Springer; 1991. [Google Scholar]
- 3.Cravatt B F, Giang D K, Mayfield S P, Boger D L, Lerner R A, Gilula N B. Nature (London) 1996;384:83–87. doi: 10.1038/384083a0. [DOI] [PubMed] [Google Scholar]
- 4.Yamada T, Palm C J, Brooks B, Kosuge T. Proc Natl Acad Sci USA. 1985;82:6522–6526. doi: 10.1073/pnas.82.19.6522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klee H, Montoya A, Horodyski F, Lichtenstein C, Garfinkel D, Fuller S, Flores C, Peschon J, Nester E, Gordon M. Proc Natl Acad Sci USA. 1984;81:1728–1732. doi: 10.1073/pnas.81.6.1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kobayashi M, Nagasawa T, Yamada H. Trends Biotechnol. 1992;10:402–408. doi: 10.1016/0167-7799(92)90283-2. [DOI] [PubMed] [Google Scholar]
- 7.Yamada H, Kobayashi M. Biosci Biotech Biochem. 1996;60:1391–1400. doi: 10.1271/bbb.60.1391. [DOI] [PubMed] [Google Scholar]
- 8.Hayashi T, Yamamoto K, Matsuo A, Otsubo K, Muramatsu S, Matsuda A, Komatsu K-I. J Ferment Bioeng. 1997;83:139–145. [Google Scholar]
- 9.Zambryski P, Tempe J, Schell J. Cell. 1989;56:193–201. doi: 10.1016/0092-8674(89)90892-1. [DOI] [PubMed] [Google Scholar]
- 10.Kobayashi M, Komeda H, Nagasawa T, Nishiyama M, Horinouchi S, Beppu T, Yamada H, Shimizu S. Eur J Biochem. 1993;217:327–336. doi: 10.1111/j.1432-1033.1993.tb18250.x. [DOI] [PubMed] [Google Scholar]
- 11.Komeda H, Kobayashi M, Shimizu S. Proc Natl Acad Sci USA. 1997;94:36–41. doi: 10.1073/pnas.94.1.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Komeda H, Kobayashi M, Shimizu S. J Biol Chem. 1996;271:15796–15802. doi: 10.1074/jbc.271.26.15796. [DOI] [PubMed] [Google Scholar]
- 13.Ettinger R A, DeLuca H F. Arch Biochem Biophys. 1995;316:14–19. doi: 10.1006/abbi.1995.1003. [DOI] [PubMed] [Google Scholar]
- 14.Ettinger R A, Ismail R, DeLuca H F. J Biol Chem. 1994;269:176–182. [PubMed] [Google Scholar]
- 15.Tsuchiya K, Fukuyama S, Kanzaki N, Kanagawa K, Negoro S, Okada H. J Bacteriol. 1989;171:3187–3191. doi: 10.1128/jb.171.6.3187-3191.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Okada H, Negoro S, Kimura H, Nakamura S. Nature (London) 1983;306:203–206. doi: 10.1038/306203a0. [DOI] [PubMed] [Google Scholar]
- 17.Mayaux J-F, Cerbelaud E, Soubrier F, Yeh P, Blanche F, Petre D. J Bacteriol. 1991;173:6694–6704. doi: 10.1128/jb.173.21.6694-6704.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ciskanik L M, Wilczek J M, Fallon R D. Appl Environ Microbiol. 1995;61:998–1003. doi: 10.1128/aem.61.3.998-1003.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hashimoto Y, Nishiyama M, Ikehata O, Horinouchi S, Beppu T. Biochim Biophys Acta. 1991;1088:225–233. doi: 10.1016/0167-4781(91)90058-t. [DOI] [PubMed] [Google Scholar]
- 20.Mayaux J-F, Cerbelaud E, Soubrier F, Faucher D, Petre D. J Bacteriol. 1990;172:6764–6773. doi: 10.1128/jb.172.12.6764-6773.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nishiyama M, Horinouchi S, Kobayashi M, Nagasawa T, Yamada H, Beppu T. J Bacteriol. 1991;173:2465–2472. doi: 10.1128/jb.173.8.2465-2472.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Silva A M, Cachau R E, Sham H L, Erickson J W. J Mol Biol. 1996;255:321–346. doi: 10.1006/jmbi.1996.0026. [DOI] [PubMed] [Google Scholar]
- 23.Beppu T. In: Recombinant DNA and Bacterial Fermentation. Thomson J A, editor. Boca Raton, FL: CRC; 1988. pp. 11–21. [Google Scholar]
- 24.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 25.Sanger F, Nicklen S, Coulson A R. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taylor J W, Ott J, Eckstein F. Nucleic Acids Res. 1985;13:8764–8785. doi: 10.1093/nar/13.24.8765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carter P, Wells J A. Nature (London) 1988;332:564–568. doi: 10.1038/332564a0. [DOI] [PubMed] [Google Scholar]
- 28.Paetzel M, Dalbey R E. Trends Biochem Sci. 1997;22:28–31. doi: 10.1016/s0968-0004(96)10065-7. [DOI] [PubMed] [Google Scholar]
- 29.Toh H, Ono M, Saigo K, Miyata T. Nature (London) 1985;315:691. [Google Scholar]
- 30.Veerapandian B, Cooper J B, Sali A, Blundell T L, Rosati R L, Dominy B W, Damon D B, Hoover D J. Protein Sci. 1992;1:322–328. doi: 10.1002/pro.5560010303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tang J, James M N G, Hsu I N, Jenkins J A, Blundell T L. Nature (London) 1978;271:618–621. doi: 10.1038/271618a0. [DOI] [PubMed] [Google Scholar]
- 32.Jakoby W B, Fredericks J. J Biol Chem. 1964;239:1978–1982. [PubMed] [Google Scholar]
- 33.Negoro S, Mitamura T, Oka K, Kanagawa K, Okada H. Eur J Biochem. 1989;185:521–524. doi: 10.1111/j.1432-1033.1989.tb15144.x. [DOI] [PubMed] [Google Scholar]
- 34.Nishiya Y, Imanaka T. J Ferment Bioeng. 1993;75:245–253. [Google Scholar]
- 35.Brammar W J, Charles I G, Matfield M, Liu C P, Drew R E, Clarke P H. FEBS Lett. 1987;215:291–294. doi: 10.1016/0014-5793(87)80164-3. [DOI] [PubMed] [Google Scholar]
- 36.Bork P, Koonin E V. Protein Sci. 1994;3:1344–1346. doi: 10.1002/pro.5560030821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Novo C, Tata R, Clemente A, Brown P R. FEBS Lett. 1995;367:275–279. doi: 10.1016/0014-5793(95)00585-w. [DOI] [PubMed] [Google Scholar]
- 38.Kobayashi M, Izui H, Nagasawa T, Yamada H. Proc Natl Acad Sci USA. 1993;90:247–251. doi: 10.1073/pnas.90.1.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Komeda H, Hori Y, Kobayashi M, Shimizu S. Proc Natl Acad Sci USA. 1996;93:10572–10577. doi: 10.1073/pnas.93.20.10572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brown D T, Turner P D, O’Reilly C. FEMS Microbiol Lett. 1995;134:143–146. doi: 10.1111/j.1574-6968.1995.tb07928.x. [DOI] [PubMed] [Google Scholar]
- 41.Wang P, Van Etten H D. Biochem Biophys Res Commun. 1992;187:1048–1054. doi: 10.1016/0006-291x(92)91303-8. [DOI] [PubMed] [Google Scholar]
- 42.Kobayashi M, Shimizu S. FEMS Microbiol Lett. 1994;120:217–224. [Google Scholar]
- 43.Nishimori K, Kawaguchi Y, Hidaka M, Uozumi T, Beppu T. J Biochem (Tokyo) 1982;91:1085–1088. doi: 10.1093/oxfordjournals.jbchem.a133758. [DOI] [PubMed] [Google Scholar]
- 44.Harris T J R, Lowe P A, Lyons A, Thomas P G, Eaton M A W, Millican T A, Patel T P, Bose C C, Carey N H, Doel M T. Nucleic Acids Res. 1982;10:2177–2187. doi: 10.1093/nar/10.7.2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin X, Wong R N S, Tang J. J Biol Chem. 1989;264:4482–4489. [PubMed] [Google Scholar]
- 46.Chang T H, Abelson J. Nucleic Acids Res. 1990;18:7180. doi: 10.1093/nar/18.23.7180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sekine M, Watanabe K, Syono K. Nucleic Acids Res. 1989;17:6400. doi: 10.1093/nar/17.15.6400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gomi K, Kitamoto K, Kumagai C. Gene. 1991;108:91–98. doi: 10.1016/0378-1119(91)90491-s. [DOI] [PubMed] [Google Scholar]
- 49.Corrick C M, Twomey A P, Hynes M J. Gene. 1987;53:63–71. doi: 10.1016/0378-1119(87)90093-x. [DOI] [PubMed] [Google Scholar]
- 50.Barrie K A, Perez E E, Lamers S L, Farmerie W G, Dunn B M, Sleasman J W, Goodenow M M. Virology. 1996;219:407–416. doi: 10.1006/viro.1996.0266. [DOI] [PubMed] [Google Scholar]