Abstract
Infection of vertebrate cells with alphaviruses normally leads to prodigious expression of virus-encoded genes and a dramatic inhibition of host protein synthesis. Recombinant Sindbis viruses and replicons have been useful as vectors for high level foreign gene expression, but the cytopathic effects of viral replication have limited their use to transient studies. We recently selected Sindbis replicons capable of persistent, noncytopathic growth in BHK cells and describe here a new generation of Sindbis vectors useful for long-term foreign gene expression based on such replicons. Foreign genes of interest as well as the dominant selectable marker puromycin N-acteyltransferase, which confers resistance to the drug puromycin, were expressed as subgenomic transcripts of noncytopathic replicons or defective-interfering genomes complemented in trans by a replicon. Based on these strategies, we developed vectors that can be initiated via either RNA or DNA transfection and analyzed them for their level and stability of foreign gene expression. Noncytopathic Sindbis vectors express reasonably high levels of protein in nearly every cell. These vectors should prove to be flexible tools for the rapid expression of heterologous genes under conditions in which cellular metabolism is not perturbed, and we illustrate their utility with a number of foreign proteins.
The ongoing molecular dissection of alphaviruses has permitted the development of alphavirus-based RNA vectors useful for transient gene expression (1). Alphaviruses infect a variety of commonly used animal cell types, replicate to high levels, and express large amounts of viral and heterologous gene products (2). Vectors based on the type alphavirus, Sindbis virus (SIN), have found use in the study of other viruses and their gene products, in cell biology, and in the delivery of foreign genes as antigens (1). Because the effects of viral replication are toxic to host cells, SIN vectors have been limited to transient expression studies (3).
The SIN genome consists of a positive-sense single-stranded RNA molecule, ≈11.7 kb in length, with a 5′-cap structure and a 3′-poly(A) tract (4). Replication proceeds through a minus-strand RNA intermediate, which is used as template for the synthesis of additional viral genomes and for the transcription of a subgenomic mRNA. This message is colinear with the 3′ one-third of the genome and encodes the SIN structural proteins. The structural genes comprise the virion components and are dispensable for viral RNA replication.
Based on a functional cDNA clone (5), several types of SIN vectors have been constructed. Viral genomes have been engineered to transcribe additional subgenomic mRNAs encoding foreign genes of interest (6, 7). RNA transfection yields recombinant virus stocks that are used to initiate foreign gene expression on infection of permissive cells. Alternatively, the SIN structural genes can simply be replaced by foreign sequences, leading to the production of RNA “replicons,” which are capable of initiating replication and gene expression after RNA transfection but are not packaged into viruses (8). The packaging of replicons is possible by supplying the viral structural genes in trans. Cotransfection of a replicon RNA and a defective viral genome that contains the cis elements for replication and structural gene expression leads to high titer stocks of packaged replicons (9).
To better understand the basis for SIN-mediated cell death, we selected for replicons with reduced cytopathic effects (I.F., E.A., T. A. Hoffman, B.M.P., M. S. Lippa, S.S., and C.M.R., unpublished data). Preliminary experiments demonstrated that the pac gene, which encodes puromycin N-acetyltransferase (PAC), could be used as a dominant selectable marker when expressed by a SIN replicon. This replicon was then used to transduce cells to puromycin resistance (purR), producing populations of cells that: (i) replicate SIN, by virtue of their ability to express PAC; and (ii) survive the process of SIN replication. Subsequent analysis demonstrated that such cells contained replicons bearing adaptive mutations, which reduced their ability to kill cells. The causal mutation was mapped for one replicon (S1) to a single amino acid change (P to L) at position 726 of the SIN nonstructural protein nsP2. The S1 replicon efficiently establishes persistent replication, albeit with reduced levels of RNA accumulation, and cells harboring the S1 replicon have normal morphology and growth rates in the presence of puromycin.
We describe here the application of the noncytopathic S1 replicon to the development of vectors for long-term expression of heterologous sequences. The resultant vectors provide a high level, long-term gene expression technology, which should prove to be useful in many areas of biological research.
MATERIALS AND METHODS
Plasmid Constructions.
All plasmids were constructed by using standard molecular biology techniques (10) and checked by restriction analysis and/or sequencing. In the interest of space, only the salient features of plasmids are described here. Details of plasmid design as well as computer readable sequence files are available on request.
pSINrep18 is the reconstructed S1 replicon, and is identical to the construction called pTSG/pacS1 (I.F., E.A., T. A. Hoffman, B.M.P., M. S. Lippa, S.S., and C.M.R., unpublished data). pSINrep19 was constructed by the addition of a second subgenomic promoter and multiple cloning sites upstream of the pac gene in pSINrep18.
p987/SINrep19/lacZ contained the SINrep19/lacZ cDNA (see below) flanked by the 5′ Rous sarcoma virus-long terminal repeat and simian virus 40 polyadenylation signals. pSINrep21 was constructed to contain the SINrep19 cDNA flanked by these same signals in a pBR322-derivative backbone.
The defective-interfering (DI) construct DI/lacZ-neo contained the lacZ gene under control of the subgenomic RNA promoter. This was followed immediately by the internal ribosome entry site element of encephalomyocarditis virus (EMC) fused to the bacterial aminoglycoside phosphotransferase gene (neo). The other DI construct, DI/luc, was described previously (11).
The multiple-cloning site was used to subclone foreign genes of interest into noncytopathic vectors. pSINrep19/lacZ contained the lacZ gene encoding the bacterial β-galactosidase (β-gal) enzyme. pSINrep19/GFP contained the green fluorescent protein (GFP) gene (12). pSINrep19/SEAP contained the secreted human placental alkaline phosphatase gene from pSEAP-basic (CLONTECH). pSINrep19/CD46 was designed to express the human CD46 molecule (gift of John P. Atkinson, Washington University). The gene encoding T7 bacteriophage (T7) RNA polymerase was subcloned from pAR1173 (13) to yield pSINrep19/T7pol.
Transcription, Transfection, and Selection of PurR Cell Populations.
Infectious transcripts were synthesized as described (14) in reactions containing 7mG(5′)ppp(5′G) (New England Biolabs). RNA (2 μg) was transfected into cells by electroporation (15). DNA was transfected either by the calcium phosphate procedure (10) or by using lipofectamine (Life Technologies, Gaithersburg, MD) according to the manufacturer’s recommendations (1 μg of DNA and 8 μl of lipid/35-mm dish). After transfection, cells were allowed to rest ≈12–18 h, and the media was changed to standard growth media containing 5 μg/ml puromycin (Sigma). Thereafter, cells were maintained in media plus puromycin and could be passaged or frozen as typical BHK cells.
Cell Staining, Enzyme Activities, and Microscopic Analyses.
Cells were stained for β-gal activity with 5-bromo-4-chloro-3-indolyl-β-galactopyranoside (X-gal) (Sigma) as described (16). Enzymatic assays for β-gal activity were performed as described (10) by using purified β-gal standards (Life Technologies). Human placental alkaline phosphatase activity present in conditioned media was analyzed by using a chemiluminescent assay (CLONTECH) according to manufacturer’s directions. Luciferase activity was measured in cell lysates by using a commercial assay according to the manufacturer’s recommendations (Promega). CD46 molecules were visualized on the surface of formaldehyde-fixed cells by incubation with rabbit anti-CD46 sera followed by secondary labeling with rhodamine-conjugated goat anti-rabbit IgG (Boehringer). GFP expression was visualized under a fluorescent microscope outfitted with a fluorescein filter.
Metabolic Labeling, Immunoprecipitation, and SDS/PAGE.
Proteins were metabolically labeled by incubating cells in methionine- and cysteine-deficient DMEM containing 2% fetal calf serum and 100 μCi/ml Expre35S35S protein-labeling mix (NEN). Proteins were harvested and immunoprecipitated according to previous methods (17, 18).
RESULTS
Noncytopathic RNA Vectors.
Noncytopathic replicon SINrep19 was designed as a double subgenomic RNA vector, using the 5′-promoter for foreign gene expression, and a 3′-promoter to drive the expression of pac (Fig. 1A). BHK cells were transfected with capped replicon RNAs synthesized in vitro, plated, and allowed to recover for 12–18 h. We then imposed selection for cells which had been productively transfected with the replicons by addition of puromycin to cell culture medium. Within 24 h, control or nontransfected cells were completely eliminated (I.F., E.A., T. A. Hoffman, B.M.P., M. S. Lippa, S.S., and C.M.R., unpublished data).
Figure 1.
Noncytopathic SIN vectors. (A) SINrep19 RNA, the double subgenomic noncytopathic RNA vector, transcribes foreign genes and the pac gene from negative-strand templates by using separate subgenomic promoters (promoter positions are indicated by arrows in both DNA and RNA constructs). (B) A bipartite noncytopathic RNA vector relies on SINrep18 to support the replication of a defective, interfering SIN genome containing a gene of interest. The 5′ terminus of the DI RNA has a tRNA Asp sequence (20). (C) SINrep21 is a DNA vector for the SINrep19 replicon. After DNA transfection, replicon RNAs were transcribed via nuclear enzymes and transported to the cytoplasm. Replicons initiate autonomous replication and express the pac gene and foreign genes of interest. Run-off refers to convenient sites for run-off transcription; MCS, multiple-cloning sites.
A comparison of total β-gal activity delivered by SINrep19/lacZ with that from other SIN/lacZ replicons is shown in Table 1. The persistent replicon produced only ≈4% of the enzyme activity produced by the cytopathic SINrep3/lacZ replicon. This is in accord with the decreased levels of genomic and subgenomic SIN RNAs observed in cells transfected with the parental noncytopathic replicon (I.F., E.A., T. A. Hoffman, B.M.P., M. S. Lippa, S.S., and C.M.R., unpublished data).
Table 1.
Expression of β-gal by different SIN replicons
| Replicon, noncytopathic/cytopathic | μg of β-gal/106 cells* |
|---|---|
| SINrep19/lacZ, noncytopathic | 1.4 ± 0.1 |
| SINrep3/lacZ, cytopathic | 40 ± 2.4 |
| SINrep18 + DI/lacZ-neo, noncytopathic | 31 ± 1.7 |
Amounts of β-gal were determined by comparison with concentration standards in enzymatic activity assays. Cell lysates were prepared following 2 days of puromycin selection (for noncytopathic replicons) or 18 h after infection at a moi of 10 (packaged cytopathic replicons). For the noncytopathic replicons, expression levels plateau after 2 days of selection; for the cytopathic replicons, 18 h precedes the onset of cytopathic effect although expression levels are only one-half maximal at this time. Standard BHK cells expressed undetectable β-gal activity. The sensitivity of this assay was ≈1 ng of purified enzyme. Values shown reflect the mean and SD for three replicate experiments.
Strategies Using DI RNAs.
A defining feature of alphavirus DI particles is increased RNA replication levels over wild-type or helper virus (19). DI genomes supported by a noncytopathic SIN replicon also could exhibit increased replication levels and, consequently, increased foreign gene production. For one SIN DI, an important determinant was found to be the substitution of a tRNA-like structure for the authentic 5′-end of a defective genome (20). DI RNAs encoding heterologous genes were constructed based on this structure and used in conjunction with a noncytopathic replicon containing a single subgenomic promoter to drive pac gene expression (SINrep18).
In one strategy, DI RNAs were transfected into purR cells already replicating SINrep18 (Fig. 1B). To phenotypically distinguish DI-transfected cells from nontransfected cells, a second dominant selectable marker was used. The neo gene, which confers resistance to the drug G418, was placed adjacent to the gene of interest and driven by the internal ribosome entry site element of encephalomyocarditis virus. The DI/lacZ-neo vector produced >20 times the level of β-gal produced by SINrep19 but slightly less than that of the cytopathic replicon (Table 1). Metabolic labeling of actinomycin-d-resistant RNA species in vivo indicated that DI RNAs replicated to higher levels than noncytopathic replicons alone and that DI/lacZ-neo-mediated expression was stable after multiple passages even in the absence of G418 selection (data not shown). These data suggest that a DI/replicon vector system is capable of high level expression of foreign genes, most likely due to increased replication of the DI genome over that of the replicon.
To further characterize the ability of noncytopathic replicons to complement a DI vector, we examined the ability of SINrep18 to induce expression of luciferase in cells constitutively transcribing DI/luc from a nuclear promoter. This transcript serves as a template for replication and transcription upon introduction of a helper virus or replicon (11). As Table 2 demonstrates, uninduced cells accumulated a low constitutive level of luciferase activity that was enhanced by infection and proportional to the amount of SIN virus added. When these cells were transfected with SINrep18 followed by puromycin selection and passaging, abundant luciferase activity accumulated in the first 5 days and was maintained for at least another 4 days. Thus, although noncytopathic replicons express less protein per unit of time than their cytopathic counterparts, their ability for prolonged expression can lead to accumulation of very high levels of protein.
Table 2.
Induction of luciferase by noncytopathic replicons or SIN virus
| Induction | RLU/106 cells* |
|---|---|
| None | 1.9 ± 0.3 × 104 |
| SINrep18, 5 days after transfection | 5.1 ± 1.7 × 107 |
| SINrep18, 9 days after transfection | 7.0 ± 0.9 × 107 |
| SIN virus, moi 0.1 | 3.3 ± 1.0 × 106 |
| SIN virus, moi 1.0 | 1.7 ± 0.5 × 107 |
Luciferase activity was determined in lysates of puromycin-selected cell populations (for noncytopathic replicons) or 8 h after infection at the indicated moi (with SIN virus, as determined by using BHK monolayers). The uninduced control reflects basal luciferase expression. Values shown reflect the mean and SD for three replicate experiments. RLU, relative light units.
Frequency of Expressing Cells and Stability of Cell Populations Upon Passaging.
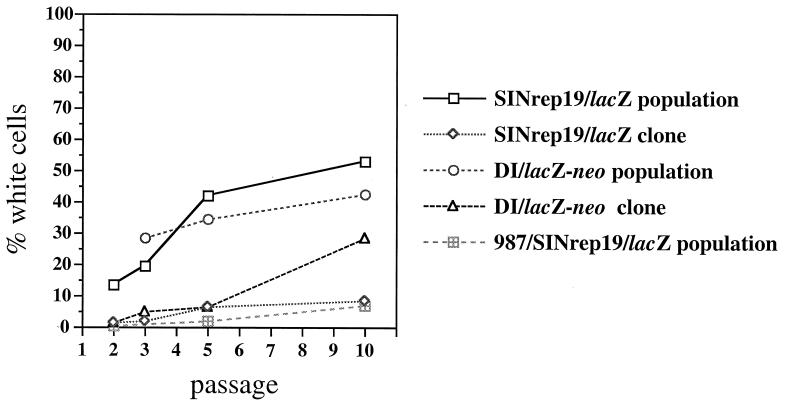
The stability of a long-term expression vector strongly defines its utility. Because RNA viruses, including SIN, are genetically quite plastic, we examined the pattern of β-gal expression via noncytopathic replicons, on a per cell basis, over time. As Fig. 2 demonstrates, a population of cells replicating SINrep19/lacZ contained >10% β-gal-negative cells within the first few passages and continued to accumulate expression-negative cells upon further passage. A similar loss of expression was seen for SINrep18 + DI/lacZ (Fig. 2) and for other marker genes (data not shown). Thus, despite continued puromycin selection, as well as the fact that the lacZ and pac genes were genetically linked, expression-negative variants arose within the selected populations. We considered two main sources for this heterogeneity: (i) replicons are likely to accumulate mutations or deletions in the lacZ gene during SIN replication; and (ii) replicons are initiated as a population, which should reflect the heterogeneity of the in vitro synthesized RNAs.
Figure 2.
Stability of β-gal expression by noncytopathic replicons. During the course of cell passage (1:10, approximately every 48 h), a portion of cells were seeded in separate wells and stained with X-gal as described in Materials and Methods. Stability of β-gal expressing cloned cell lines was assessed by using clones selected for high level expression. The percentage of β-gal-negative cells was calculated by microscopic examination of several random fields.
To address the first of these concerns. we examined the stability of expression in cloned populations of β-gal-positive cells. As seen in Fig. 2, a clonal derivative of SINrep19/lacZ maintained >90% β-gal expression through 10 passages; similar results were seen with multiple cell clones (data not shown). The stability of SINrep18 + DI/lacZ also was improved upon cloning a β-gal-positive cell, although the percent of expression dropped by the 10th passage. For cloned populations of both SINrep19/lacZ and SINrep18 + DI/lacZ, omitting selection had no effect on the percentage of β-gal-expressing cells over seven passages (data not shown). These results suggest that the loss of expression does occur as a result of SIN replication, but this does not fully account for the instability in uncloned populations. In a practical sense, this instability can be managed through the use of early passage cells and by directly cloning expression-positive cells.
The high mutation rate of SP6 RNA polymerase may contribute to the initial heterogeneity within a replicon population. In vivo transcription of functional RNA replicons by using the host cell’s nuclear RNA polymerase II could be an alternative to in vitro transcription, similar to published strategies (21, 22). This approach was first tested by constructing p987/SINrep19/lacZ, which contained the SINrep19/lacZ cDNA flanked by the Rous sarcoma virus 5′-long terminal repeat promoter and the simian virus 40 polyadenylation signal. Replication of this replicon was initiated by transient transfection of plasmid DNA and puromycin selection of transfected populations. Such populations exhibited prolonged expression of β-gal in >90% of cells, although a small but detectable loss of expression was observed (Fig. 2). No differences in expression level were noticed between SINrep19- vs. p987/SINrep19-expressed products. Based on this design, a general DNA vector, pSINrep21, was constructed in a medium copy vector (Fig. 1C).
Expression of Various Heterologous Proteins.
Some representative genes expressed by using noncytopathic replicons are shown in Fig. 3. SINrep19/GFP contains a synthetic human codon-optimized version of the Aequeorea victoria GFP gene (12), which led to bright fluorescence of purR populations (Fig. 3A). Fig. 3B demonstrates the pattern of X-gal staining for a fifth passage SINrep19/lacZ-expressing cell population.
Figure 3.
Expression of model proteins via noncytopathic vectors. Cells were selected after transfection with SINrep19/GFP (A), SINrep19/lacZ (B), SINrep19/CD46 (C and E), or SINrep18 (D and F). All of the cells were representative populations selected for purR as described in the text. Cells were prepared by X-gal staining (B) or immunofluorescent staining for CD46 (C and D). (E and F) show the condition of cells 54 h after infection with MV [multiplicity of infection (moi) 0.1].
We were particularly interested in using noncytopathic replicons to deliver proteins to the secretory pathway because one consequence of SIN infection may be the disregulation of these cellular compartments (23). We expressed the human cell surface glycoprotein CD46, which is involved in complement regulation and is the receptor for measles virus (MV). CD46 was expressed on the surface of cells replicating SINrep19/CD46 (Fig. 3 C and D). As this receptor is required for MV binding and entry, we tested whether CD46 expression could confer MV permissivity to BHK cells. As seen in Fig. 3 E and F, a population of SINrep19/CD46 cells demonstrated the syncytia formation typically induced by MV infection, whereas SINrep18 cells remained nonpermissive.
In addition, a nonmembrane anchored form of human placental alkaline phosphatase (SEAP) was secreted into the medium of cells replicating SINrep19/SEAP at a rate of 400 ng/106 cells/24 h. Similarly, a truncated form of the E2 glycoprotein of hepatitis C virus (HCV), lacking a transmembrane anchor, was properly glycosylated and targeted for secretion (data not shown). The yellow fever (YF) virus structural proteins prM/E also have been expressed via SINrep19 and are properly processed into prM and E (data not shown). These data show that noncytopathic SIN vectors can deliver biologically active proteins to the secretory pathway.
Expression of T7 RNA Polymerase.
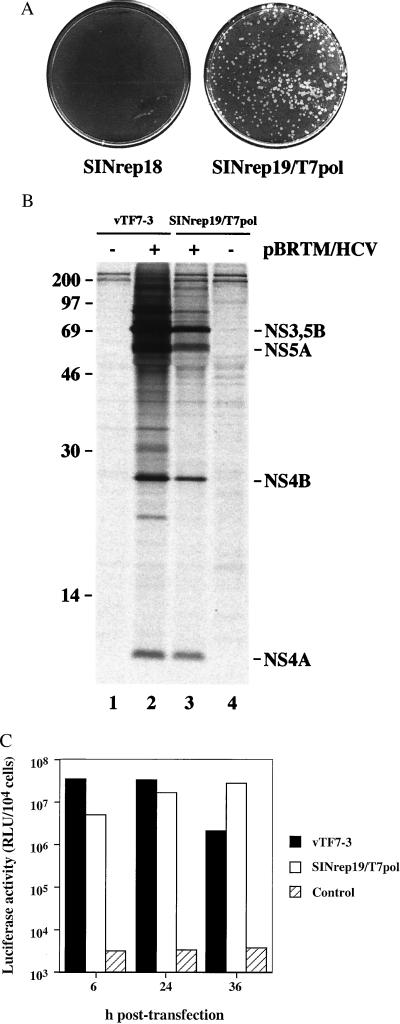
We examined the ability of noncytopathic replicons to express phage T7 RNA polymerase directly to host cells. This enzyme is commonly delivered to mammalian cells by using a recombinant vaccinia virus, vTF7–3, and it has found broad utility in transient overexpression of foreign genes cloned downstream of the T7 promoter and appropriate translational signals (24). The function of T7 RNA polymerase expressed via SINrep19/T7pol was demonstrated by using various reporters. In one set of experiments, we transfected these cells with plasmid DNA containing a functional Mengo virus clone under transcriptional control of the T7 promoter. As can be seen in Fig. 4A, the formation of Mengo virus plaques was specific to cells harboring SINrep19/T7pol. These plaques were identical in size and morphology to Mengo virus initiated from in vitro transcripts (data not shown).
Figure 4.
Functional expression of T7 RNA polymerase. (A) Mengo virus plaques after transfection of control BHK cells (SINrep18) or T7-expressing BHK cells (SINrep19/T7pol) with a T7-driven functional clone of Mengo virus. (B) Expression of the HCV polyprotein with (lanes 2 and 3) or without (lanes 1 and 4) transfection of pBRTM/HCV827–3011. Lanes 1 and 2 were derived from cells infected with vTF7–3, whereas lanes 3 and 4 represent cells containing SINrep19/T7pol. (C) Expression of luciferase after transfection of pEMCLucβgAn into cells expressing T7 RNA polymerase via vTF7–3, SINrep19/T7pol, or not expressing T7 RNA polymerase (control). Lysates were prepared at 6, 24, or 36 h after transfection. This experiment is representative of three independent experiments.
In another set of experiments, T7 RNA polymerase-expressing cells were transfected with pBRTM/HCV827–3011, a plasmid containing the nonstructural region of HCV under control of the T7 promoter (25). The expected products were immunoprecipitated, indicating that the HCV polyprotein was expressed and properly processed (Fig. 4B, lanes 3 and 4). For comparison, we also expressed the phage polymerase via vTF7–3, which yielded the same products but in greater abundance (Fig. 4B, lanes 1 and 2).
To better quantitate the amount of T7-expressed product, we examined the expression of luciferase over time in cells transfected with a plasmid encoding the luc gene driven by a T7 promoter, pEMCLucβgAn (a gift of Jon A. Wolff, University of Wisconsin). Cells expressing T7 RNA polymerase via vTF7–3 or SINrep19/T7pol had abundant luciferase activity at all times examined. Vaccinia-infected cells demonstrated an earlier peak of activity, which waned by 36 h, consistent with the destruction of infected cells by vaccinia. In contrast, SINrep19/T7pol continued to accumulate luciferase, surpassing the activity in vTF7–3-infected cells between 24 and 36 h. These data show that SINrep19/T7pol can express functional T7 RNA polymerase at a level comparable with that of vTF7–3 but with less cytotoxicity.
DISCUSSION
Noncytopathic SIN replicons have been applied to the design of vectors for long-term foreign gene expression. Populations of expressing cells are derived by RNA or DNA transfection of the appropriate vector and selection of replicon-bearing cells with puromycin. Advantages of this system include the ease and rapidity of obtaining foreign gene-expressing populations and the reasonably high level of foreign gene expression. Although not yet characterized as an expression system, a selectable noncytopathic subgenomic RNA replicon based on the flavivirus Kunjin has been recently described (26).
Based on our data, the quantity of foreign gene expression appears to be a function of SIN RNA replication levels. SINrep19/lacZ expressed 1–2 μg of β-gal/106 cells, which corresponds to ≈107 molecules of enzyme/cell. In contrast, SINrep3/lacZ produced ≈2 × 108 molecules/cell. By comparison, conventional Rous sarcoma virus promoter-enhancer driven constructs express ≈ 5 × 106 molecules/cell (27). Physiological levels of the src-family kinases p56lck and p59fyn are ≈105/cell (28); those of the abundant extracellular signal-related kinases Erk1 and Erk2 have been estimated at 106/cell (29). Thus despite their apparent inefficiency when compared with cytopathic SIN vectors, noncytopathic replicons are capable of delivering physiologically relevant levels of foreign gene expression. In applications for which more robust long-term expression is desired, the replicon/DI system may be more useful. The SIN cis-acting translational enhancer element (30, 31) was tested and found to have no enhancing effect on foreign gene expression in the context of the noncytopathic vectors (data not shown). Conversely, for even greater attenuation of foreign gene expression, it should be possible to make use of variant SIN junction regions, which are less efficient subgenomic mRNA promoters (7).
Although stability issues remain a concern, nearly every cell in a puromycin-selected population expresses the foreign gene product. A likely source of expression-negative cells is the accumulation of mutations arising during SIN RNA replication. These changes presumably affect foreign gene expression directly because mutations affecting SIN replication or expression of PAC would be selected against. Although the error frequency of the SIN replicase is unknown, RNA viruses are notoriously error-prone, with estimates (32, 33) as high as 10−3-10−4. Thus, the noncytopathic replicons may be most useful for expression studies in early passages after selection. This is most easily accomplished by constructing a seed-lot system for a given cell population. In addition, clonal derivatives of RNA vector-bearing cells show a marked increase in stability.
A more dramatic improvement in expression stability was seen by using replicons initiated by DNA transfection. Scant biochemical data exist on the error frequency of the DNA-dependent RNA polymerases used in this study. However, T7 RNA polymerase is estimated to have an error rate between 10−4 and 10−5, consisting mostly of 1-nt insertions and deletions (34, 35). Eukaryotic RNA polymerase II (wheat germ) has an estimated error frequency of ≈5 × 10−6 on synthetic polynucleotide substrates (36). In addition, eukaryotic RNA polymerase II has additional mechanisms to insure even more accurate transcription (37). Based on the available evidence, we surmise that pSINrep21 generates transcripts of superior fidelity to that of pSINrep19, initially seeding more homogenous cell populations. We have observed however that some foreign genes do not express well via pSINrep21. Several attempts to generate a cell population expressing T7 RNA polymerase via pSINrep21/T7pol failed (data not shown). Given that the identical construct in pSINrep19 was functional, there must have been a gene-specific defect. Perhaps the T7 RNA polymerase gene contains a cryptic splice site, which disrupts the reading frame when expressed via the nucleus. Nevertheless, all other genes that have been tested were compatible with pSINrep21.
A limitation of noncytopathic replicons is the cell type restriction. SINrep19 and SINrep21 both work in all derivatives of BHK-21 and Chinese hamster ovary cells that we have tested (data not shown). Vero cells seem to work less efficiently, but replicon-bearing cells do survive selection. Cell types that have failed to support the noncytopathic replicons include primary chicken embryo fibroblasts, MDBK, MDCK, 293, HeLa, and PC12 cells. These results are surprising because all are permissive for SIN replication after infection or RNA transfection. Although untested, it is possible that differences in replication efficiency or in interferon production/responsiveness play a role in determining the host range of the noncytopathic SIN replicons. In any case, BHK, Chinese hamster ovary, and Vero cells are widely used in the fields of cell biology and virology, suggesting that noncytopathic vectors will be useful in these areas. We are currently investigating the possibility of deriving pan-adaptive replicons with increased host range. If such obstacles can be overcome, perhaps noncytopathic vectors may find additional application in vivo, such as in delivering therapeutic genes or foreign antigens (38).
The noncytopathic replicons described here permit the rapid generation of cell lines expressing nontoxic gene products for many applications. We have illustrated their use with a number of reporters, including β-gal, SEAP, and GFP; and several genes with relevance to virology: CD46, a truncated HCV E2, and YF prM/E. In addition, we recently have shown that noncytopathic replicons can be used to deliver YF NS1 for trans-complementation of a YF genome lacking NS1 function (39). The apparent compatibility of noncytopathic vectors with infection by a number of heterologous viruses (YF, Mengo, MV, and all others tested), further demonstrates the utility of these vectors for future studies in virology.
The ability to express T7 RNA polymerase with a noncytopathic replicon may be a useful alternative to other methods of in vivo T7 transcription such as with the recombinant vaccinia virus, vTF7–3. The popularity of this latter approach has led to the construction of numerous T7-driven vectors, and it appears that they can be used directly with SINrep19/T7pol (Fig. 4 B and C). This may be most useful where the cytopathic effects of vaccinia infection interfere with cellular functions under study. Additionally, we anticipate that this system may be useful for reverse genetics of the negative-strand RNA viruses, which currently use in vivo T7 transcripts for the generation of infectious RNA and viral nucleoproteins (40, 41).
In summary, we have applied our knowledge of SIN biology to the development of noncytopathic vectors, which are capable of medium to high level expression of foreign genes in nearly every transfected cell. These vectors should find use in a variety of biological systems.
Acknowledgments
We thank Carol Read and Rebecca Moran for expert technical assistance. Special thanks are extended to John P. Atkinson, Ann Palmenberg, John Majors, Bernard Moss, Brian Seed, and Jon A. Wolff for sharing plasmid DNAs. We also are grateful to many colleagues for helpful discussions during the course of this work and to Sean Amberg, Karen Reed, and Paul Olivo for critical reading of the manuscript. This work was supported by grants from the Public Health Service (AI24134 and AI11377). B.M.P. is a Visiting Professor on leave from the Albert Szent-Györgyi Medical University, Department of Microbiology, Szeged, Hungary, and is supported in part by a grant from the Hungarian Science Foundation OTKA (T26040/98).
ABBREVIATIONS
- DI
defective-interfering
- PAC
puromycin N-acetyltransferase
- purR
puromycin resistance
- T7
bacteriophage T7
- GFP
green fluorescent protein
- HCV
hepatitis C virus
- SEAP
secreted alkaline phosphatase
- SIN
Sindbis virus
- MV
measles virus
- YF
yellow fever virus
- moi
multiplicity of infection
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
A Commentary on this article begins on page 12750.
References
- 1. Frolov I, Hoffman T A, Prágai B M, Dryga S A, Huang H V, Schlesinger S, Rice C M. Proc Natl Acad Sci USA. 1996;93:11371–11377. doi: 10.1073/pnas.93.21.11371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Strauss J H, Strauss E G. Microbiol Rev. 1994;58:491–562. doi: 10.1128/mr.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frolov I, Schlesinger S. J Virol. 1994;68:1721–1727. doi: 10.1128/jvi.68.3.1721-1727.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Strauss E G, Rice C M, Strauss J H. Virology. 1984;133:92–110. doi: 10.1016/0042-6822(84)90428-8. [DOI] [PubMed] [Google Scholar]
- 5.Rice C M, Levis R, Strauss J H, Huang H V. J Virol. 1987;61:3809–3819. doi: 10.1128/jvi.61.12.3809-3819.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hahn C S, Hahn Y S, Braciale T J, Rice C M. Proc Natl Acad Sci USA. 1992;89:2679–2683. doi: 10.1073/pnas.89.7.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raju R, Huang H V. J Virol. 1991;65:2501–2510. doi: 10.1128/jvi.65.5.2501-2510.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiong C, Levis R, Shen P, Schlesinger S, Rice C, Huang H V. Science. 1989;243:1188–1191. doi: 10.1126/science.2922607. [DOI] [PubMed] [Google Scholar]
- 9.Bredenbeek P J, Frolov I, Rice C M, Schlesinger S. J Virol. 1993;67:6439–6446. doi: 10.1128/jvi.67.11.6439-6446.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. New York: Greene & Wiley; 1993. [Google Scholar]
- 11.Olivo P D, Frolov I, Schlesinger S. Virology. 1994;198:381–384. doi: 10.1006/viro.1994.1046. [DOI] [PubMed] [Google Scholar]
- 12.Haas J, Park E-U, Seed B. Curr Biol. 1996;6:315–324. doi: 10.1016/s0960-9822(02)00482-7. [DOI] [PubMed] [Google Scholar]
- 13.Davanloo P, Rosenberg A H, Dunn J J, Studier F W. Proc Natl Acad Sci USA. 1984;81:2035–2039. doi: 10.1073/pnas.81.7.2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levis R, Weiss B G, Tsiang M, Huang H, Schlesinger S. Cell. 1986;44:137–145. doi: 10.1016/0092-8674(86)90492-7. [DOI] [PubMed] [Google Scholar]
- 15.Liljeström P, Lusa S, Huylebroeck D, Garoff H. J Virol. 1991;65:4107–4113. doi: 10.1128/jvi.65.8.4107-4113.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lim K, Chae C-B. BioTechniques. 1989;7:576–579. [PubMed] [Google Scholar]
- 17.Chambers T J, McCourt D W, Rice C M. Virology. 1990;177:159–174. doi: 10.1016/0042-6822(90)90470-c. [DOI] [PubMed] [Google Scholar]
- 18.Lin C, Lindenbach B D, Prágai B, McCourt D W, Rice C M. J Virol. 1994;68:5063–5073. doi: 10.1128/jvi.68.8.5063-5073.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schlesinger S, Schlesinger M J. In: Replication of Togaviridae and Flaviviridae. Fields B N, Knipe D M, editors. Vol. 1. New York: Lippincott-Raven; 1990. pp. 697–711. [Google Scholar]
- 20.Monroe S S, Schlesinger S. Proc Natl Acad Sci USA. 1983;80:3279–3283. doi: 10.1073/pnas.80.11.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dubensky T W, Jr, Driver D A, Polo J M, Belli B A, Latham E M, Ibanez C E, Chada S, Brumm D, Banks T A, Mento S J, et al. J Virol. 1996;70:508–519. doi: 10.1128/jvi.70.1.508-519.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herweijer H, Latendresse J S, Williams P, Zhang G, Danko I, Schlesinger S, Wolff J A. Hum Gene Ther. 1995;6:1161–1167. doi: 10.1089/hum.1995.6.9-1161. [DOI] [PubMed] [Google Scholar]
- 23.Pugachev K V, Mason P W, Frey T K. Virology. 1995;209:155–166. doi: 10.1006/viro.1995.1239. [DOI] [PubMed] [Google Scholar]
- 24.Moss B, Elroy-Stein O, Mizukami T, Alexander W A, Fuerst T R. Nature (London) 1990;348:91–92. doi: 10.1038/348091a0. [DOI] [PubMed] [Google Scholar]
- 25.Grakoui A, Wychowski C, Lin C, Feinstone S M, Rice C M. J Virol. 1993;67:1385–1395. doi: 10.1128/jvi.67.3.1385-1395.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khromykh A A, Westaway E G. J Virol. 1997;71:1497–1505. doi: 10.1128/jvi.71.2.1497-1505.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang H V, Rice C M, Xiong C, Schlesinger S. Virus Genes. 1989;3:85–91. doi: 10.1007/BF00301989. [DOI] [PubMed] [Google Scholar]
- 28.Olszowy M W, Leuchtmann P L, Veillette A, Shaw A S. J Immunol. 1995;155:4236–4240. [PubMed] [Google Scholar]
- 29.Ferrell J E., Jr Trends Biochem Sci. 1996;21:460–466. doi: 10.1016/s0968-0004(96)20026-x. [DOI] [PubMed] [Google Scholar]
- 30.Frolov I, Schlesinger S. J Virol. 1994;68:8111–8117. doi: 10.1128/jvi.68.12.8111-8117.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Frolov I, Schlesinger S. J Virol. 1996;70:1182–1190. doi: 10.1128/jvi.70.2.1182-1190.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Steinhauer D A, Holland J J. J Virol. 1986;57:219–228. doi: 10.1128/jvi.57.1.219-228.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ward C D, Flanegan J B. J Virol. 1988;62:558–562. doi: 10.1128/jvi.62.2.558-562.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boyer J C, Bebenek K, Kunkel T A. Proc Natl Acad Sci USA. 1992;89:6919–6923. doi: 10.1073/pnas.89.15.6919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sooknanan R, Howes M, Read L, Malek L T. BioTechniques. 1994;17:1077–1085. [PubMed] [Google Scholar]
- 36.De Mercoyrol L, Corda Y, Job C, Job D. Eur J Biochem. 1992;206:49–58. doi: 10.1111/j.1432-1033.1992.tb16900.x. [DOI] [PubMed] [Google Scholar]
- 37.Thomas M J, Platas A A, Hawley D K. Cell. 1998;93:627–637. doi: 10.1016/s0092-8674(00)81191-5. [DOI] [PubMed] [Google Scholar]
- 38.Hariharan M J, Driver D A, Townsend K, Brumm D, Polo J M, Belli B A, Catton D J, Hsu D, Mittelstaedt D, McCormack J E, et al. J Virol. 1998;72:950–958. doi: 10.1128/jvi.72.2.950-958.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lindenbach B D, Rice C M. J Virol. 1997;71:9608–9617. doi: 10.1128/jvi.71.12.9608-9617.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Conzelmann K K. J Gen Virol. 1996;77:381–389. doi: 10.1099/0022-1317-77-3-381. [DOI] [PubMed] [Google Scholar]
- 41.Palese P, Zheng H, Engelhardt O G, Pleschka S, Garcia-Sastre A. Proc Natl Acad Sci USA. 1996;93:11354–11358. doi: 10.1073/pnas.93.21.11354. [DOI] [PMC free article] [PubMed] [Google Scholar]