Abstract
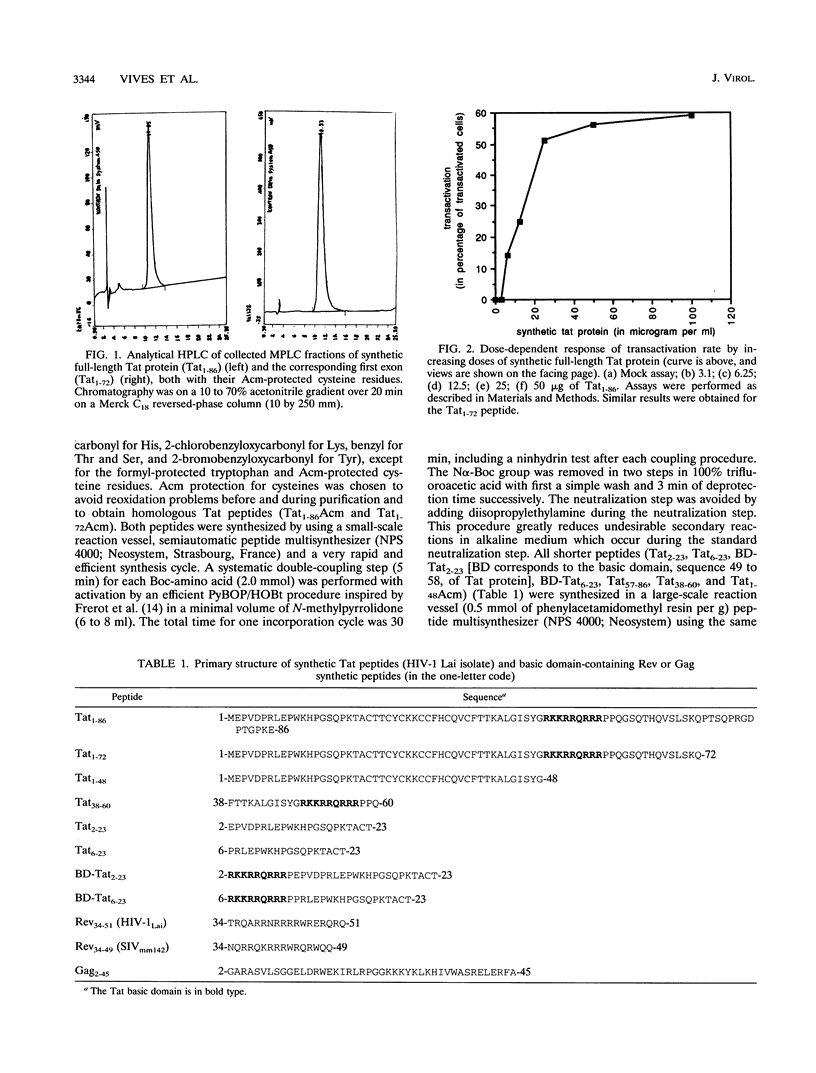
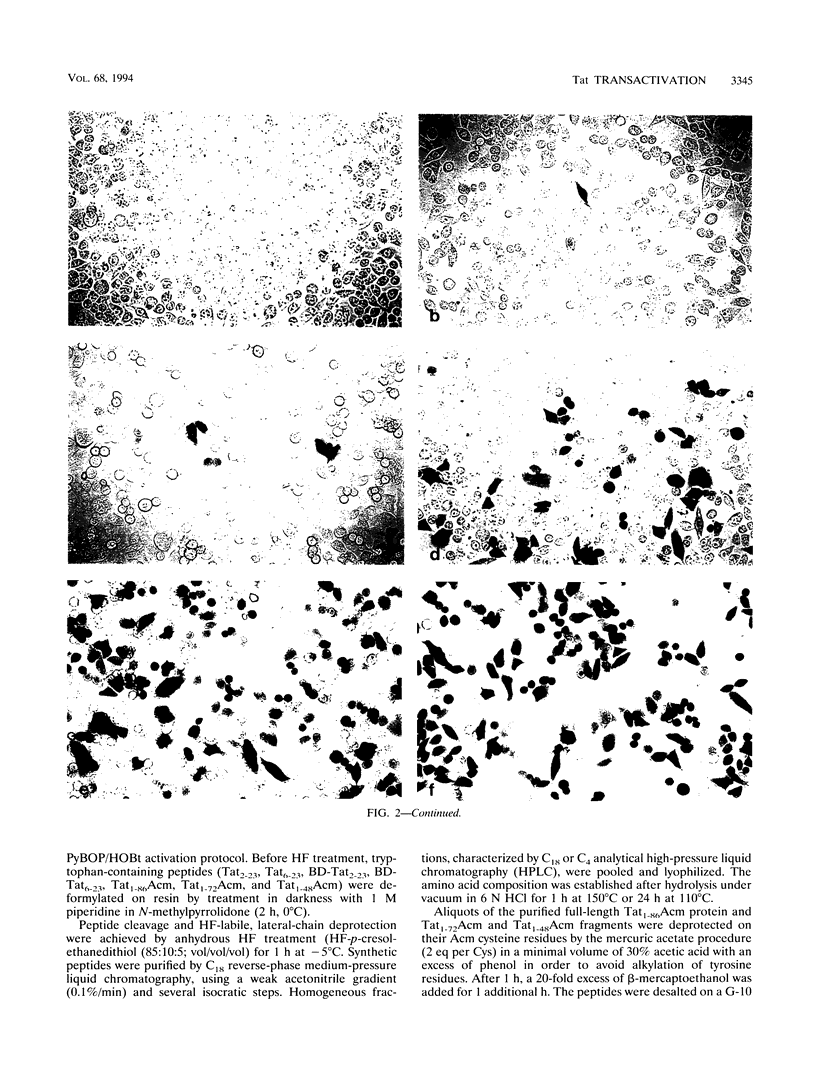
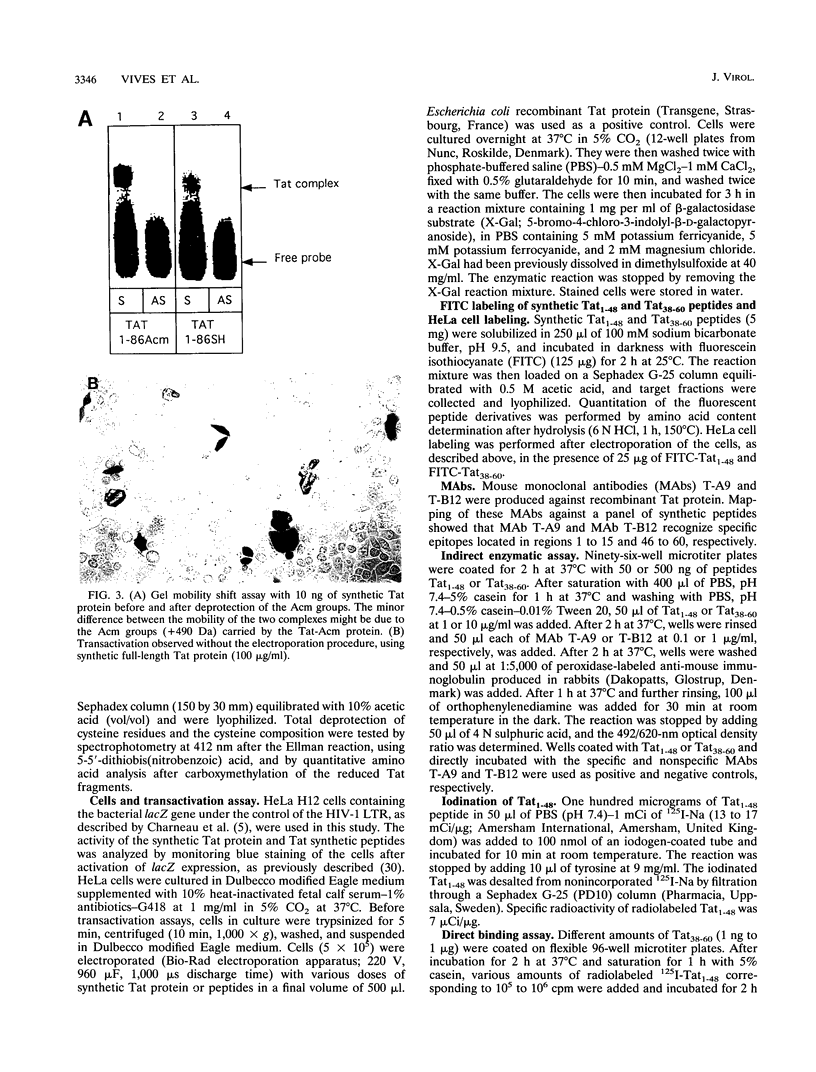
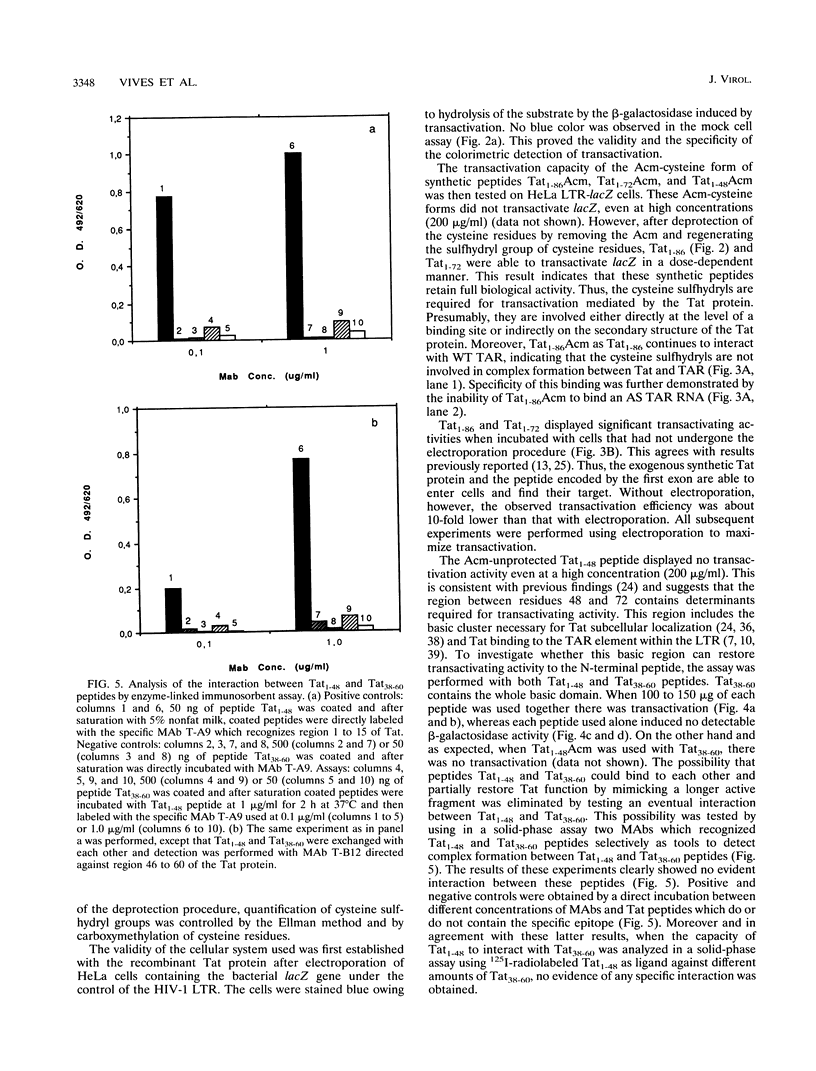
To study the structure relationship of different Tat domains, the full-length Tat protein Tat1-86, the gene product of the first exon Tat1-72 which retains full activity of the protein, and a panel of shorter peptides mimicking different regions of the primary structure of the Tat protein were chemically synthesized by the solid-phase method, using an efficient protocol. Synthetic Tat1-86 and Tat1-72 transactivated beta-galactosidase activity in HeLa cells containing the lacZ gene under the control of the human immunodeficiency virus type 1 long terminal repeat. Analyses of the activity of Tat1-86 and Tat1-72 with the sulfhydryl of cysteine residues free or protected by the acetamidomethyl group showed that only the Tat fragments with deprotected cysteine residues retain transactivation ability. In contrast, peptide Tat1-48 was inactive, with cysteine residues either free or protected. Similarly, other shorter synthetic peptides covering the different Tat domains were inactive. Interestingly, when peptides Tat1-48 and Tat38-60 were used simultaneously, a significant transactivation was obtained. This result suggests that both peptide domains are implicated in transactivation, probably by acting at two different sites. This permits us to propose a fundamentally new step in the understanding of the molecular mechanism of Tat transactivation.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anfinsen C. B. Principles that govern the folding of protein chains. Science. 1973 Jul 20;181(4096):223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- Arya S. K., Guo C., Josephs S. F., Wong-Staal F. Trans-activator gene of human T-lymphotropic virus type III (HTLV-III). Science. 1985 Jul 5;229(4708):69–73. doi: 10.1126/science.2990040. [DOI] [PubMed] [Google Scholar]
- Barry P. A., Pratt-Lowe E., Unger R. E., Luciw P. A. Cellular factors regulate transactivation of human immunodeficiency virus type 1. J Virol. 1991 Mar;65(3):1392–1399. doi: 10.1128/jvi.65.3.1392-1399.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braddock M., Chambers A., Wilson W., Esnouf M. P., Adams S. E., Kingsman A. J., Kingsman S. M. HIV-1 TAT "activates" presynthesized RNA in the nucleus. Cell. 1989 Jul 28;58(2):269–279. doi: 10.1016/0092-8674(89)90841-6. [DOI] [PubMed] [Google Scholar]
- Charneau P., Alizon M., Clavel F. A second origin of DNA plus-strand synthesis is required for optimal human immunodeficiency virus replication. J Virol. 1992 May;66(5):2814–2820. doi: 10.1128/jvi.66.5.2814-2820.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez L. G., Jr, Scheraga H. A. Intrinsic stabilities of portions of the ribonuclease molecule. Biochemistry. 1980 Mar 4;19(5):1005–1012. doi: 10.1021/bi00546a027. [DOI] [PubMed] [Google Scholar]
- Chun R., Glabe C. G., Fan H. Chemical synthesis of biologically active tat trans-activating protein of human immunodeficiency virus type 1. J Virol. 1990 Jun;64(6):3074–3077. doi: 10.1128/jvi.64.6.3074-3077.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordingley M. G., LaFemina R. L., Callahan P. L., Condra J. H., Sardana V. V., Graham D. J., Nguyen T. M., LeGrow K., Gotlib L., Schlabach A. J. Sequence-specific interaction of Tat protein and Tat peptides with the transactivation-responsive sequence element of human immunodeficiency virus type 1 in vitro. Proc Natl Acad Sci U S A. 1990 Nov;87(22):8985–8989. doi: 10.1073/pnas.87.22.8985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen B. R. The HIV-1 Tat protein: an RNA sequence-specific processivity factor? Cell. 1990 Nov 16;63(4):655–657. doi: 10.1016/0092-8674(90)90129-3. [DOI] [PubMed] [Google Scholar]
- Desai K., Loewenstein P. M., Green M. Isolation of a cellular protein that binds to the human immunodeficiency virus Tat protein and can potentiate transactivation of the viral promoter. Proc Natl Acad Sci U S A. 1991 Oct 15;88(20):8875–8879. doi: 10.1073/pnas.88.20.8875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingwall C., Ernberg I., Gait M. J., Green S. M., Heaphy S., Karn J., Lowe A. D., Singh M., Skinner M. A., Valerio R. Human immunodeficiency virus 1 tat protein binds trans-activation-responsive region (TAR) RNA in vitro. Proc Natl Acad Sci U S A. 1989 Sep;86(18):6925–6929. doi: 10.1073/pnas.86.18.6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel A. D., Biancalana S., Hudson D. Activity of synthetic peptides from the Tat protein of human immunodeficiency virus type 1. Proc Natl Acad Sci U S A. 1989 Oct;86(19):7397–7401. doi: 10.1073/pnas.86.19.7397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel A. D., Bredt D. S., Pabo C. O. Tat protein from human immunodeficiency virus forms a metal-linked dimer. Science. 1988 Apr 1;240(4848):70–73. doi: 10.1126/science.2832944. [DOI] [PubMed] [Google Scholar]
- Frankel A. D., Pabo C. O. Cellular uptake of the tat protein from human immunodeficiency virus. Cell. 1988 Dec 23;55(6):1189–1193. doi: 10.1016/0092-8674(88)90263-2. [DOI] [PubMed] [Google Scholar]
- Furie B., Schechter A. N., Sachs D. H., Anfinsen C. B. An immunological approach to the conformational equilibrium of staphylococcal nuclease. J Mol Biol. 1975 Mar 15;92(4):497–506. doi: 10.1016/0022-2836(75)90305-8. [DOI] [PubMed] [Google Scholar]
- Gatignol A., Buckler-White A., Berkhout B., Jeang K. T. Characterization of a human TAR RNA-binding protein that activates the HIV-1 LTR. Science. 1991 Mar 29;251(5001):1597–1600. doi: 10.1126/science.2011739. [DOI] [PubMed] [Google Scholar]
- Gatignol A., Kumar A., Rabson A., Jeang K. T. Identification of cellular proteins that bind to the human immunodeficiency virus type 1 trans-activation-responsive TAR element RNA. Proc Natl Acad Sci U S A. 1989 Oct;86(20):7828–7832. doi: 10.1073/pnas.86.20.7828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaynor R., Soultanakis E., Kuwabara M., Garcia J., Sigman D. S. Specific binding of a HeLa cell nuclear protein to RNA sequences in the human immunodeficiency virus transactivating region. Proc Natl Acad Sci U S A. 1989 Jul;86(13):4858–4862. doi: 10.1073/pnas.86.13.4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green M., Loewenstein P. M. Autonomous functional domains of chemically synthesized human immunodeficiency virus tat trans-activator protein. Cell. 1988 Dec 23;55(6):1179–1188. doi: 10.1016/0092-8674(88)90262-0. [DOI] [PubMed] [Google Scholar]
- Hauber J., Malim M. H., Cullen B. R. Mutational analysis of the conserved basic domain of human immunodeficiency virus tat protein. J Virol. 1989 Mar;63(3):1181–1187. doi: 10.1128/jvi.63.3.1181-1187.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurrell J. G., Smith J. A., Leach S. J. Immunological measurements of conformational motility in regions of the myoglobin molecule. Biochemistry. 1977 Jan 25;16(2):175–185. doi: 10.1021/bi00621a003. [DOI] [PubMed] [Google Scholar]
- Jeyapaul J., Reddy M. R., Khan S. A. Activity of synthetic tat peptides in human immunodeficiency virus type 1 long terminal repeat-promoted transcription in a cell-free system. Proc Natl Acad Sci U S A. 1990 Sep;87(18):7030–7034. doi: 10.1073/pnas.87.18.7030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeyapaul J., Seshamma T., Khan S. A. Synthetic HIV-1 Tat can dissociate HeLa nuclear protein-TAR RNA complexes in vitro: a novel Tat-nuclear protein interaction. Oncogene. 1991 Sep;6(9):1507–1513. [PubMed] [Google Scholar]
- Kamine J., Chinnadurai G. Synergistic activation of the human immunodeficiency virus type 1 promoter by the viral Tat protein and cellular transcription factor Sp1. J Virol. 1992 Jun;66(6):3932–3936. doi: 10.1128/jvi.66.6.3932-3936.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H., Sumimoto H., Pognonec P., Chen C. H., Rosen C. A., Roeder R. G. HIV-1 Tat acts as a processivity factor in vitro in conjunction with cellular elongation factors. Genes Dev. 1992 Apr;6(4):655–666. doi: 10.1101/gad.6.4.655. [DOI] [PubMed] [Google Scholar]
- Krippl B., Ferguson B., Jones N., Rosenberg M., Westphal H. Mapping of functional domains in adenovirus E1A proteins. Proc Natl Acad Sci U S A. 1985 Nov;82(22):7480–7484. doi: 10.1073/pnas.82.22.7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann D. A., Frankel A. D. Endocytosis and targeting of exogenous HIV-1 Tat protein. EMBO J. 1991 Jul;10(7):1733–1739. doi: 10.1002/j.1460-2075.1991.tb07697.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marciniak R. A., Garcia-Blanco M. A., Sharp P. A. Identification and characterization of a HeLa nuclear protein that specifically binds to the trans-activation-response (TAR) element of human immunodeficiency virus. Proc Natl Acad Sci U S A. 1990 May;87(9):3624–3628. doi: 10.1073/pnas.87.9.3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy J. A., Meinwald Y. C., Scheraga H. A. Immunochemical determination of conformational equilibria for fragments of the A alpha chain of fibrinogen. Biochemistry. 1982 Apr 13;21(8):1794–1806. doi: 10.1021/bi00537a015. [DOI] [PubMed] [Google Scholar]
- Rappaport J., Lee S. J., Khalili K., Wong-Staal F. The acidic amino-terminal region of the HIV-1 Tat protein constitutes an essential activating domain. New Biol. 1989 Oct;1(1):101–110. [PubMed] [Google Scholar]
- Rice A. P., Carlotti F. Mutational analysis of the conserved cysteine-rich region of the human immunodeficiency virus type 1 Tat protein. J Virol. 1990 Apr;64(4):1864–1868. doi: 10.1128/jvi.64.4.1864-1868.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice A. P., Chan F. Tat protein of human immunodeficiency virus type 1 is a monomer when expressed in mammalian cells. Virology. 1991 Nov;185(1):451–454. doi: 10.1016/0042-6822(91)90797-f. [DOI] [PubMed] [Google Scholar]
- Rocancourt D., Bonnerot C., Jouin H., Emerman M., Nicolas J. F. Activation of a beta-galactosidase recombinant provirus: application to titration of human immunodeficiency virus (HIV) and HIV-infected cells. J Virol. 1990 Jun;64(6):2660–2668. doi: 10.1128/jvi.64.6.2660-2668.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen C. A., Sodroski J. G., Haseltine W. A. The location of cis-acting regulatory sequences in the human T cell lymphotropic virus type III (HTLV-III/LAV) long terminal repeat. Cell. 1985 Jul;41(3):813–823. doi: 10.1016/s0092-8674(85)80062-3. [DOI] [PubMed] [Google Scholar]
- Rosen C. A., Terwilliger E., Dayton A., Sodroski J. G., Haseltine W. A. Intragenic cis-acting art gene-responsive sequences of the human immunodeficiency virus. Proc Natl Acad Sci U S A. 1988 Apr;85(7):2071–2075. doi: 10.1073/pnas.85.7.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruben S., Perkins A., Purcell R., Joung K., Sia R., Burghoff R., Haseltine W. A., Rosen C. A. Structural and functional characterization of human immunodeficiency virus tat protein. J Virol. 1989 Jan;63(1):1–8. doi: 10.1128/jvi.63.1.1-8.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs D. H., Schechter A. N., Eastlake A., Anfinsen C. B. Antibodies to a distinct antigenic determinant of staphylococcal nuclease. J Immunol. 1972 Dec;109(6):1300–1310. [PubMed] [Google Scholar]
- Seigel L. J., Ratner L., Josephs S. F., Derse D., Feinberg M. B., Reyes G. R., O'Brien S. J., Wong-Staal F. Transactivation induced by human T-lymphotropic virus type III (HTLV III) maps to a viral sequence encoding 58 amino acids and lacks tissue specificity. Virology. 1986 Jan 15;148(1):226–231. doi: 10.1016/0042-6822(86)90419-8. [DOI] [PubMed] [Google Scholar]
- Sheline C. T., Milocco L. H., Jones K. A. Two distinct nuclear transcription factors recognize loop and bulge residues of the HIV-1 TAR RNA hairpin. Genes Dev. 1991 Dec;5(12B):2508–2520. doi: 10.1101/gad.5.12b.2508. [DOI] [PubMed] [Google Scholar]
- Siomi H., Shida H., Maki M., Hatanaka M. Effects of a highly basic region of human immunodeficiency virus Tat protein on nucleolar localization. J Virol. 1990 Apr;64(4):1803–1807. doi: 10.1128/jvi.64.4.1803-1807.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodroski J., Rosen C., Wong-Staal F., Salahuddin S. Z., Popovic M., Arya S., Gallo R. C., Haseltine W. A. Trans-acting transcriptional regulation of human T-cell leukemia virus type III long terminal repeat. Science. 1985 Jan 11;227(4683):171–173. doi: 10.1126/science.2981427. [DOI] [PubMed] [Google Scholar]
- Subramanian T., Kuppuswamy M., Venkatesh L., Srinivasan A., Chinnadurai G. Functional substitution of the basic domain of the HIV-1 trans-activator, Tat, with the basic domain of the functionally heterologous Rev. Virology. 1990 May;176(1):178–183. doi: 10.1016/0042-6822(90)90242-j. [DOI] [PubMed] [Google Scholar]
- Weeks K. M., Ampe C., Schultz S. C., Steitz T. A., Crothers D. M. Fragments of the HIV-1 Tat protein specifically bind TAR RNA. Science. 1990 Sep 14;249(4974):1281–1285. doi: 10.1126/science.2205002. [DOI] [PubMed] [Google Scholar]
- Wright C. M., Felber B. K., Paskalis H., Pavlakis G. N. Expression and characterization of the trans-activator of HTLV-III/LAV virus. Science. 1986 Nov 21;234(4779):988–992. doi: 10.1126/science.3490693. [DOI] [PubMed] [Google Scholar]