Abstract
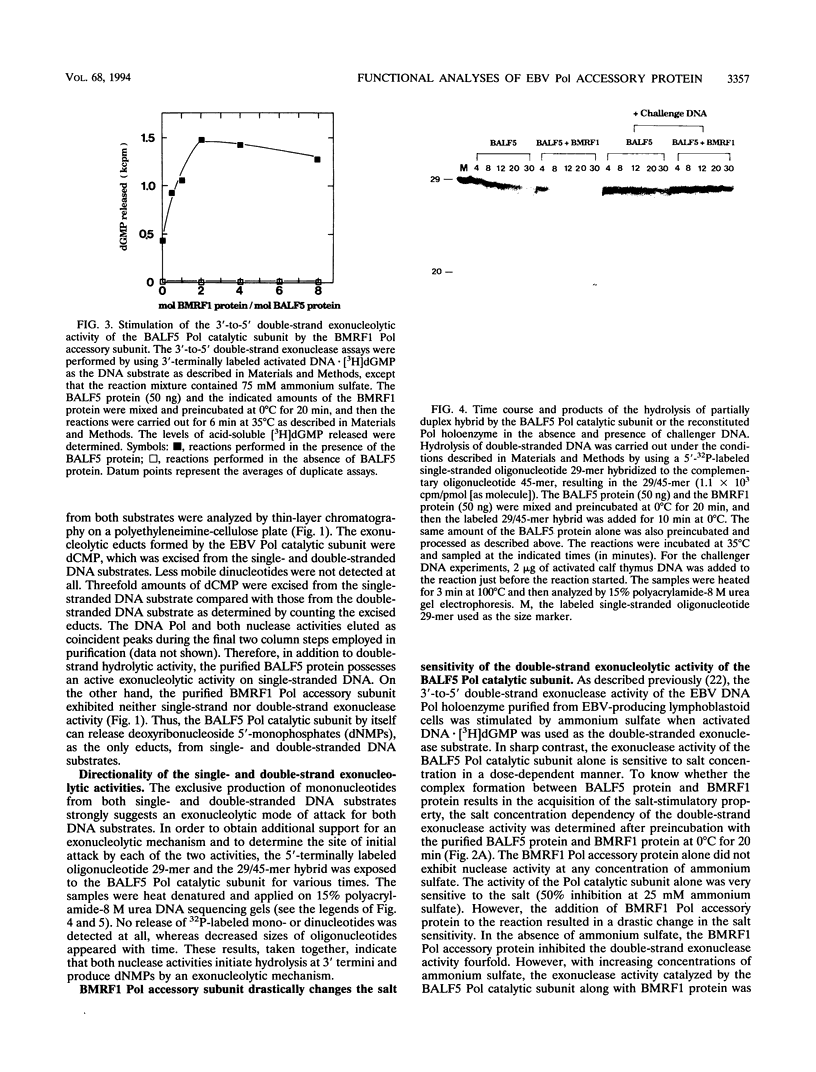
The Epstein-Barr virus (EBV) DNA polymerase catalytic subunit, BALF5 gene product, possesses an intrinsic 3'-to 5' proofreading exonuclease activity in addition to 5'-to-3' DNA polymerase activity (T. Tsurumi, A. Kobayashi, K. Tamai, T. Daikoku, R. Kurachi, and Y. Nishiyama, J. Virol. 67:4651-4658, 1993). The exonuclease hydrolyzed both double-and single-stranded DNA substrates with 3'-to-5' directionality, releasing deoxyribonucleoside 5'-monophosphates. The double-strand exonucleolytic activity catalyzed by the BALF5 polymerase catalytic subunit was very sensitive to high ionic strength, whereas the single-strand exonucleolytic activity was moderately resistant. The addition of the BMRF1 polymerase accessory subunit to the reaction enhanced the double-strand exonucleolytic activity in the presence of high concentrations of ammonium sulfate (fourfold stimulation at 75 mM ammonium sulfate). Optimal stimulation was obtained when the molar ratio of BMRF1 protein to BALF5 protein was 2 and higher, identical to the values required for reconstituting the optimum DNA polymerizing activity (T. Tsurumi, T. Daikoku, R. Kurachi, and Y. Nishiyama, J. Virol. 67:7648-7653, 1993). Furthermore, product size analyses revealed that the polymerase catalytic subunit alone excised a few nucleotides from the 3' termini of the primer hybridized to template DNA and that the addition of the BMFR1 polymerase accessory subunit stimulated the nucleotide excision several times. In contrast, the hydrolysis of single-stranded DNA by the BALF5 protein was not affected by the addition of the BMRF1 polymerase accessory subunit at all. These observations suggest that the BMRF1 polymerase accessory subunit forms a complex with the BALF5 polymerase catalytic subunit to stabilize the interaction of the holoenzyme complex with the 3'-OH end of the primer on the template DNA during exonucleolysis. On the other hand, challenger DNA experiments revealed that the BALF5 polymerase catalytic subunit alone stably binds to the primer terminus in a stationary state, whereas the reconstituted polymerase holoenzyme is unstable. The instability of the initiation complex of the EBV DNA polymerase would allow the rapid removal of the EBV DNA polymerase holoenzyme from the lagging strand after it has replicated up to the previous Okazaki fragment. This feature of the EBV DNA polymerase holoenzyme in a stationary state is in marked contrast to the moving holoenzyme complex tightly bound to the primer end during polymerization and exonucleolysis.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baer R., Bankier A. T., Biggin M. D., Deininger P. L., Farrell P. J., Gibson T. J., Hatfull G., Hudson G. S., Satchwell S. C., Séguin C. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature. 1984 Jul 19;310(5974):207–211. doi: 10.1038/310207a0. [DOI] [PubMed] [Google Scholar]
- Bedinger P., Alberts B. M. The 3'-5' proofreading exonuclease of bacteriophage T4 DNA polymerase is stimulated by other T4 DNA replication proteins. J Biol Chem. 1983 Aug 25;258(16):9649–9656. [PubMed] [Google Scholar]
- Brutlag D., Kornberg A. Enzymatic synthesis of deoxyribonucleic acid. 36. A proofreading function for the 3' leads to 5' exonuclease activity in deoxyribonucleic acid polymerases. J Biol Chem. 1972 Jan 10;247(1):241–248. [PubMed] [Google Scholar]
- Chiou J. F., Li J. K., Cheng Y. C. Demonstration of a stimulatory protein for virus-specified DNA polymerase in phorbol ester-treated Epstein-Barr virus-carrying cells. Proc Natl Acad Sci U S A. 1985 Sep;82(17):5728–5731. doi: 10.1073/pnas.82.17.5728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fixman E. D., Hayward G. S., Hayward S. D. trans-acting requirements for replication of Epstein-Barr virus ori-Lyt. J Virol. 1992 Aug;66(8):5030–5039. doi: 10.1128/jvi.66.8.5030-5039.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerschmidt W., Sugden B. Identification and characterization of oriLyt, a lytic origin of DNA replication of Epstein-Barr virus. Cell. 1988 Nov 4;55(3):427–433. doi: 10.1016/0092-8674(88)90028-1. [DOI] [PubMed] [Google Scholar]
- Hori K., Mark D. F., Richardson C. C. Deoxyribonucleic acid polymerase of bacteriophage T7. Characterization of the exonuclease activities of the gene 5 protein and the reconstituted polymerase. J Biol Chem. 1979 Nov 25;254(22):11598–11604. [PubMed] [Google Scholar]
- Huang W. M., Lehman I. R. On the exonuclease activity of phage T4 deoxyribonucleic acid polymerase. J Biol Chem. 1972 May 25;247(10):3139–3146. [PubMed] [Google Scholar]
- Jarvis T. C., Paul L. S., Hockensmith J. W., von Hippel P. H. Structural and enzymatic studies of the T4 DNA replication system. II. ATPase properties of the polymerase accessory protein complex. J Biol Chem. 1989 Jul 25;264(21):12717–12729. [PubMed] [Google Scholar]
- Kallin B., Sternås L., Saemundssen A. K., Luka J., Jörnvall H., Eriksson B., Tao P. Z., Nilsson M. T., Klein G. Purification of Epstein-Barr virus DNA polymerase from P3HR-1 cells. J Virol. 1985 May;54(2):561–568. doi: 10.1128/jvi.54.2.561-568.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiehl A., Dorsky D. I. Cooperation of EBV DNA polymerase and EA-D(BMRF1) in vitro and colocalization in nuclei of infected cells. Virology. 1991 Sep;184(1):330–340. doi: 10.1016/0042-6822(91)90849-7. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. Exonucleolytic proofreading. Cell. 1988 Jun 17;53(6):837–840. doi: 10.1016/s0092-8674(88)90189-4. [DOI] [PubMed] [Google Scholar]
- Li J. S., Zhou B. S., Dutschman G. E., Grill S. P., Tan R. S., Cheng Y. C. Association of Epstein-Barr virus early antigen diffuse component and virus-specified DNA polymerase activity. J Virol. 1987 Sep;61(9):2947–2949. doi: 10.1128/jvi.61.9.2947-2949.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munn M. M., Alberts B. M. DNA footprinting studies of the complex formed by the T4 DNA polymerase holoenzyme at a primer-template junction. J Biol Chem. 1991 Oct 25;266(30):20034–20044. [PubMed] [Google Scholar]
- Tsurumi T. Characterization of 3'-to 5'-exonuclease activity associated with Epstein-Barr virus DNA polymerase. Virology. 1991 May;182(1):376–381. doi: 10.1016/0042-6822(91)90685-5. [DOI] [PubMed] [Google Scholar]
- Tsurumi T., Daikoku T., Kurachi R., Nishiyama Y. Functional interaction between Epstein-Barr virus DNA polymerase catalytic subunit and its accessory subunit in vitro. J Virol. 1993 Dec;67(12):7648–7653. doi: 10.1128/jvi.67.12.7648-7653.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsurumi T., Kobayashi A., Tamai K., Daikoku T., Kurachi R., Nishiyama Y. Functional expression and characterization of the Epstein-Barr virus DNA polymerase catalytic subunit. J Virol. 1993 Aug;67(8):4651–4658. doi: 10.1128/jvi.67.8.4651-4658.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsurumi T. Primer terminus recognition and highly processive replication by Epstein-Barr virus DNA polymerase. Biochem J. 1991 Dec 15;280(Pt 3):703–708. doi: 10.1042/bj2800703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsurumi T. Purification and characterization of the DNA-binding activity of the Epstein-Barr virus DNA polymerase accessory protein BMRF1 gene products, as expressed in insect cells by using the baculovirus system. J Virol. 1993 Mar;67(3):1681–1687. doi: 10.1128/jvi.67.3.1681-1687.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsurumi T. Selective inhibition of the 3'-to-5' exonuclease activity associated with Epstein-Barr virus DNA polymerase by ribonucleoside 5'-monophosphates. Virology. 1992 Aug;189(2):803–807. doi: 10.1016/0042-6822(92)90611-r. [DOI] [PubMed] [Google Scholar]
- Venkatesan M., Nossal N. G. Bacteriophage T4 gene 44/62 and gene 45 polymerase accessory proteins stimulate hydrolysis of duplex DNA by T4 DNA polymerase. J Biol Chem. 1982 Oct 25;257(20):12435–12443. [PubMed] [Google Scholar]
- Yates J. L., Guan N. Epstein-Barr virus-derived plasmids replicate only once per cell cycle and are not amplified after entry into cells. J Virol. 1991 Jan;65(1):483–488. doi: 10.1128/jvi.65.1.483-488.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates J., Warren N., Reisman D., Sugden B. A cis-acting element from the Epstein-Barr viral genome that permits stable replication of recombinant plasmids in latently infected cells. Proc Natl Acad Sci U S A. 1984 Jun;81(12):3806–3810. doi: 10.1073/pnas.81.12.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young M. C., Reddy M. K., von Hippel P. H. Structure and function of the bacteriophage T4 DNA polymerase holoenzyme. Biochemistry. 1992 Sep 22;31(37):8675–8690. doi: 10.1021/bi00152a001. [DOI] [PubMed] [Google Scholar]