Abstract
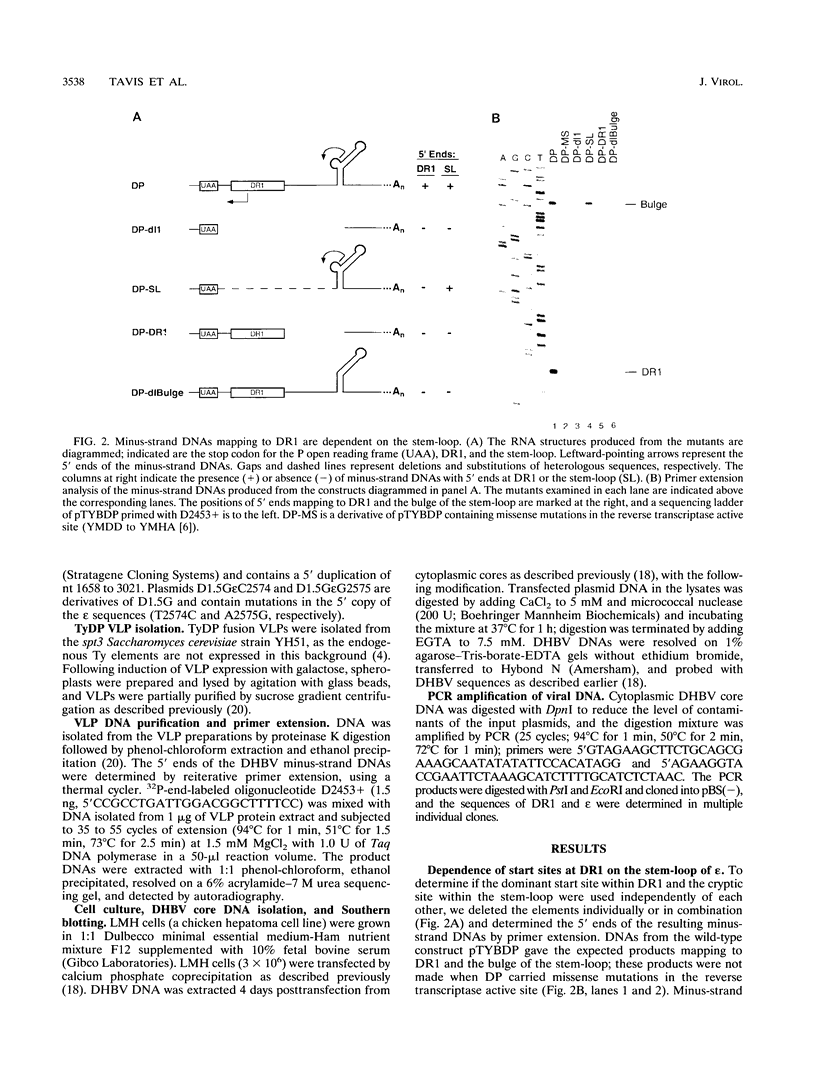
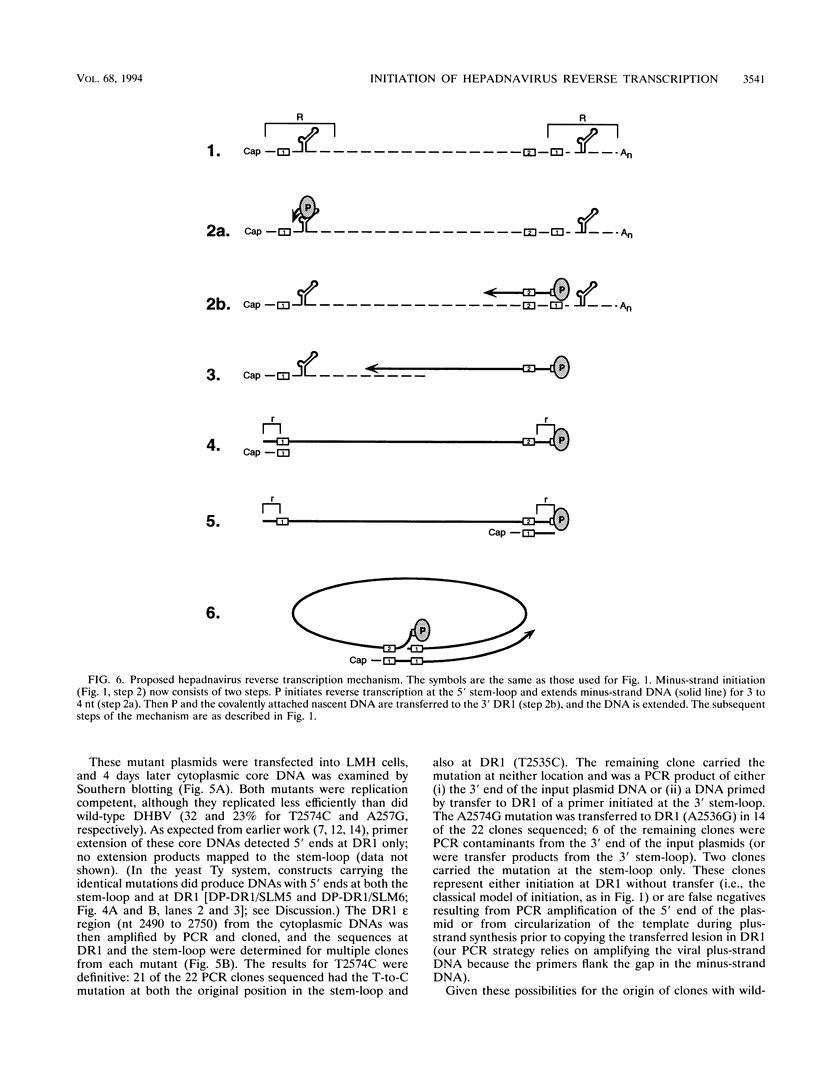
Replication of the hepadnavirus genome occurs by reverse transcription of an RNA pregenome and is mediated by the viral polymerase; the polymerase is also required for packaging of the pregenome through interaction with the RNA packaging signal, epsilon. Previous work suggested that reverse transcription of minus-strand DNA initiates within the sequence element DR1 (direct repeat 1) and that disruption of DR1 activates a cryptic initiation site in a downstream copy of epsilon. However, using active duck hepatitis B virus polymerase expressed in a yeast Ty vector system, we demonstrate that synthesis of minus-strand DNAs with 5' ends at DR1 requires the stem-loop of epsilon, whereas the production of DNAs mapping to epsilon does not require DR1. Mutations at epsilon that remove homology between epsilon and DR1 eliminate reverse transcripts with 5' ends in DR1, and restoring homology at DR1 to a mutant epsilon partially restores DNAs mapping to DR1. Insertions of one nucleotide into the bulge region of the epsilon stem-loop increase the length of minus-strand DNA whose 5' ends map to DR1 by one nucleotide. Thus, very short minus-strand primers are initiated within epsilon, rather than in DR1 as previously supposed; they are then transferred to a four-nucleotide homology in DR1. Transfer was also observed in vivo during replication of duck hepatitis B virus in avian cells; in this case, transfer is from the 5' copy of epsilon to the 3' copy of DR1. This minus-strand transfer reaction is likely to be a general feature of all hepadnaviruses.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ayola B., Kanda P., Lanford R. E. High level expression and phosphorylation of hepatitis B virus polymerase in insect cells with recombinant baculoviruses. Virology. 1993 May;194(1):370–373. doi: 10.1006/viro.1993.1270. [DOI] [PubMed] [Google Scholar]
- Bartenschlager R., Junker-Niepmann M., Schaller H. The P gene product of hepatitis B virus is required as a structural component for genomic RNA encapsidation. J Virol. 1990 Nov;64(11):5324–5332. doi: 10.1128/jvi.64.11.5324-5332.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartenschlager R., Schaller H. The amino-terminal domain of the hepadnaviral P-gene encodes the terminal protein (genome-linked protein) believed to prime reverse transcription. EMBO J. 1988 Dec 20;7(13):4185–4192. doi: 10.1002/j.1460-2075.1988.tb03315.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke J. D., Styles C. A., Fink G. R. Saccharomyces cerevisiae SPT3 gene is required for transposition and transpositional recombination of chromosomal Ty elements. Mol Cell Biol. 1986 Nov;6(11):3575–3581. doi: 10.1128/mcb.6.11.3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch V., Bartenschlager R., Radziwill G., Schaller H. The duck hepatitis B virus P-gene codes for protein strongly associated with the 5'-end of the viral DNA minus strand. Virology. 1988 Oct;166(2):475–485. doi: 10.1016/0042-6822(88)90518-1. [DOI] [PubMed] [Google Scholar]
- Chang L. J., Hirsch R. C., Ganem D., Varmus H. E. Effects of insertional and point mutations on the functions of the duck hepatitis B virus polymerase. J Virol. 1990 Nov;64(11):5553–5558. doi: 10.1128/jvi.64.11.5553-5558.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condreay L. D., Wu T. T., Aldrich C. E., Delaney M. A., Summers J., Seeger C., Mason W. S. Replication of DHBV genomes with mutations at the sites of initiation of minus- and plus-strand DNA synthesis. Virology. 1992 May;188(1):208–216. doi: 10.1016/0042-6822(92)90751-a. [DOI] [PubMed] [Google Scholar]
- Ganem D., Varmus H. E. The molecular biology of the hepatitis B viruses. Annu Rev Biochem. 1987;56:651–693. doi: 10.1146/annurev.bi.56.070187.003251. [DOI] [PubMed] [Google Scholar]
- Hirsch R. C., Lavine J. E., Chang L. J., Varmus H. E., Ganem D. Polymerase gene products of hepatitis B viruses are required for genomic RNA packaging as wel as for reverse transcription. Nature. 1990 Apr 5;344(6266):552–555. doi: 10.1038/344552a0. [DOI] [PubMed] [Google Scholar]
- Junker-Niepmann M., Bartenschlager R., Schaller H. A short cis-acting sequence is required for hepatitis B virus pregenome encapsidation and sufficient for packaging of foreign RNA. EMBO J. 1990 Oct;9(10):3389–3396. doi: 10.1002/j.1460-2075.1990.tb07540.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lien J. M., Petcu D. J., Aldrich C. E., Mason W. S. Initiation and termination of duck hepatitis B virus DNA synthesis during virus maturation. J Virol. 1987 Dec;61(12):3832–3840. doi: 10.1128/jvi.61.12.3832-3840.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar-Kimber K. L., Summers J. W., Mason W. S. Mapping of the cohesive overlap of duck hepatitis B virus DNA and of the site of initiation of reverse transcription. J Virol. 1984 Jul;51(1):181–191. doi: 10.1128/jvi.51.1.181-191.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollack J. R., Ganem D. An RNA stem-loop structure directs hepatitis B virus genomic RNA encapsidation. J Virol. 1993 Jun;67(6):3254–3263. doi: 10.1128/jvi.67.6.3254-3263.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radziwill G., Zentgraf H., Schaller H., Bosch V. The duck hepatitis B virus DNA polymerase is tightly associated with the viral core structure and unable to switch to an exogenous template. Virology. 1988 Mar;163(1):123–132. doi: 10.1016/0042-6822(88)90239-5. [DOI] [PubMed] [Google Scholar]
- Seeger C., Maragos J. Identification and characterization of the woodchuck hepatitis virus origin of DNA replication. J Virol. 1990 Jan;64(1):16–23. doi: 10.1128/jvi.64.1.16-23.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staprans S., Loeb D. D., Ganem D. Mutations affecting hepadnavirus plus-strand DNA synthesis dissociate primer cleavage from translocation and reveal the origin of linear viral DNA. J Virol. 1991 Mar;65(3):1255–1262. doi: 10.1128/jvi.65.3.1255-1262.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers J., Mason W. S. Replication of the genome of a hepatitis B--like virus by reverse transcription of an RNA intermediate. Cell. 1982 Jun;29(2):403–415. doi: 10.1016/0092-8674(82)90157-x. [DOI] [PubMed] [Google Scholar]
- Tavis J. E., Ganem D. Expression of functional hepatitis B virus polymerase in yeast reveals it to be the sole viral protein required for correct initiation of reverse transcription. Proc Natl Acad Sci U S A. 1993 May 1;90(9):4107–4111. doi: 10.1073/pnas.90.9.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G. H., Seeger C. Novel mechanism for reverse transcription in hepatitis B viruses. J Virol. 1993 Nov;67(11):6507–6512. doi: 10.1128/jvi.67.11.6507-6512.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G. H., Seeger C. The reverse transcriptase of hepatitis B virus acts as a protein primer for viral DNA synthesis. Cell. 1992 Nov 13;71(4):663–670. doi: 10.1016/0092-8674(92)90599-8. [DOI] [PubMed] [Google Scholar]







