Abstract
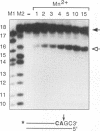
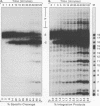
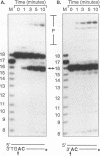
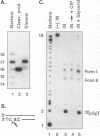
Although integration generally is considered a critical step in the retrovirus life cycle, it has been reported that visna virus, which causes degenerative neurologic disease in sheep, can productively infect sheep choroid plexus cells without detectable integration. To ascertain whether the integrase (IN) of visna virus is an inherently defective enzyme and to create tools for further study of integration of the phylogenetically related human immunodeficiency virus type 1 (HIV-1), we purified visna virus IN by using a bacterial expression system and applied various in vitro oligonucleotide-based assays to studying this protein. We found that visna virus IN demonstrates the full repertoire of in vitro functions characteristic of retroviral integrases. In particular, visna virus IN exhibits site-specific endonuclease activity following the invariant CA found two nucleotides from the 3' ends of viral DNA (processing activity), joins processed oligonucleotides to various sites on other oligonucleotides (strand transfer or integration activity), and reverses the integration reaction by resolving a complex that mimics one end of viral DNA integrated into host DNA (disintegration activity). In addition, although it has been reported that purified HIV-1 IN cannot specifically nick visna virus DNA ends, purified visna virus IN does specifically process and integrate HIV-1 DNA ends.
Full text
PDF











Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andrésson O. S., Elser J. E., Tobin G. J., Greenwood J. D., Gonda M. A., Georgsson G., Andrésdóttir V., Benediktsdóttir E., Carlsdóttir H. M., Mäntylä E. O. Nucleotide sequence and biological properties of a pathogenic proviral molecular clone of neurovirulent visna virus. Virology. 1993 Mar;193(1):89–105. doi: 10.1006/viro.1993.1106. [DOI] [PubMed] [Google Scholar]
- Bowerman B., Brown P. O., Bishop J. M., Varmus H. E. A nucleoprotein complex mediates the integration of retroviral DNA. Genes Dev. 1989 Apr;3(4):469–478. doi: 10.1101/gad.3.4.469. [DOI] [PubMed] [Google Scholar]
- Braun M. J., Clements J. E., Gonda M. A. The visna virus genome: evidence for a hypervariable site in the env gene and sequence homology among lentivirus envelope proteins. J Virol. 1987 Dec;61(12):4046–4054. doi: 10.1128/jvi.61.12.4046-4054.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown P. O., Bowerman B., Varmus H. E., Bishop J. M. Retroviral integration: structure of the initial covalent product and its precursor, and a role for the viral IN protein. Proc Natl Acad Sci U S A. 1989 Apr;86(8):2525–2529. doi: 10.1073/pnas.86.8.2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushman F. D., Craigie R. Activities of human immunodeficiency virus (HIV) integration protein in vitro: specific cleavage and integration of HIV DNA. Proc Natl Acad Sci U S A. 1991 Feb 15;88(4):1339–1343. doi: 10.1073/pnas.88.4.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushman F. D., Engelman A., Palmer I., Wingfield P., Craigie R. Domains of the integrase protein of human immunodeficiency virus type 1 responsible for polynucleotidyl transfer and zinc binding. Proc Natl Acad Sci U S A. 1993 Apr 15;90(8):3428–3432. doi: 10.1073/pnas.90.8.3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow S. A., Vincent K. A., Ellison V., Brown P. O. Reversal of integration and DNA splicing mediated by integrase of human immunodeficiency virus. Science. 1992 Feb 7;255(5045):723–726. doi: 10.1126/science.1738845. [DOI] [PubMed] [Google Scholar]
- Cobrinik D., Aiyar A., Ge Z., Katzman M., Huang H., Leis J. Overlapping retrovirus U5 sequence elements are required for efficient integration and initiation of reverse transcription. J Virol. 1991 Jul;65(7):3864–3872. doi: 10.1128/jvi.65.7.3864-3872.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craigie R., Fujiwara T., Bushman F. The IN protein of Moloney murine leukemia virus processes the viral DNA ends and accomplishes their integration in vitro. Cell. 1990 Aug 24;62(4):829–837. doi: 10.1016/0092-8674(90)90126-y. [DOI] [PubMed] [Google Scholar]
- Drelich M., Haenggi M., Mous J. Conserved residues Pro-109 and Asp-116 are required for interaction of the human immunodeficiency virus type 1 integrase protein with its viral DNA substrate. J Virol. 1993 Aug;67(8):5041–5044. doi: 10.1128/jvi.67.8.5041-5044.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drelich M., Wilhelm R., Mous J. Identification of amino acid residues critical for endonuclease and integration activities of HIV-1 IN protein in vitro. Virology. 1992 Jun;188(2):459–468. doi: 10.1016/0042-6822(92)90499-f. [DOI] [PubMed] [Google Scholar]
- Engelman A., Bushman F. D., Craigie R. Identification of discrete functional domains of HIV-1 integrase and their organization within an active multimeric complex. EMBO J. 1993 Aug;12(8):3269–3275. doi: 10.1002/j.1460-2075.1993.tb05996.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman A., Craigie R. Identification of conserved amino acid residues critical for human immunodeficiency virus type 1 integrase function in vitro. J Virol. 1992 Nov;66(11):6361–6369. doi: 10.1128/jvi.66.11.6361-6369.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman A., Mizuuchi K., Craigie R. HIV-1 DNA integration: mechanism of viral DNA cleavage and DNA strand transfer. Cell. 1991 Dec 20;67(6):1211–1221. doi: 10.1016/0092-8674(91)90297-c. [DOI] [PubMed] [Google Scholar]
- Fitzgerald M. L., Vora A. C., Zeh W. G., Grandgenett D. P. Concerted integration of viral DNA termini by purified avian myeloblastosis virus integrase. J Virol. 1992 Nov;66(11):6257–6263. doi: 10.1128/jvi.66.11.6257-6263.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T., Mizuuchi K. Retroviral DNA integration: structure of an integration intermediate. Cell. 1988 Aug 12;54(4):497–504. doi: 10.1016/0092-8674(88)90071-2. [DOI] [PubMed] [Google Scholar]
- Harris J. D., Blum H., Scott J., Traynor B., Ventura P., Haase A. Slow virus visna: reproduction in vitro of virus from extrachromosomal DNA. Proc Natl Acad Sci U S A. 1984 Nov;81(22):7212–7215. doi: 10.1073/pnas.81.22.7212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris J. D., Scott J. V., Traynor B., Brahic M., Stowring L., Ventura P., Haase A. T., Peluso R. Visna virus DNA: discovery of a novel gapped structure. Virology. 1981 Sep;113(2):573–583. doi: 10.1016/0042-6822(81)90185-9. [DOI] [PubMed] [Google Scholar]
- Hong T., Murphy E., Groarke J., Drlica K. Human immunodeficiency virus type 1 DNA integration: fine structure target analysis using synthetic oligonucleotides. J Virol. 1993 Feb;67(2):1127–1131. doi: 10.1128/jvi.67.2.1127-1131.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones K. S., Coleman J., Merkel G. W., Laue T. M., Skalka A. M. Retroviral integrase functions as a multimer and can turn over catalytically. J Biol Chem. 1992 Aug 15;267(23):16037–16040. [PubMed] [Google Scholar]
- Jonsson C. B., Donzella G. A., Roth M. J. Characterization of the forward and reverse integration reactions of the Moloney murine leukemia virus integrase protein purified from Escherichia coli. J Biol Chem. 1993 Jan 15;268(2):1462–1469. [PubMed] [Google Scholar]
- Jonsson C. B., Roth M. J. Role of the His-Cys finger of Moloney murine leukemia virus integrase protein in integration and disintegration. J Virol. 1993 Sep;67(9):5562–5571. doi: 10.1128/jvi.67.9.5562-5571.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz R. A., Mack J. P., Merkel G., Kulkosky J., Ge Z., Leis J., Skalka A. M. Requirement for a conserved serine in both processing and joining activities of retroviral integrase. Proc Natl Acad Sci U S A. 1992 Aug 1;89(15):6741–6745. doi: 10.1073/pnas.89.15.6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz R. A., Merkel G., Kulkosky J., Leis J., Skalka A. M. The avian retroviral IN protein is both necessary and sufficient for integrative recombination in vitro. Cell. 1990 Oct 5;63(1):87–95. doi: 10.1016/0092-8674(90)90290-u. [DOI] [PubMed] [Google Scholar]
- Katzman M., Katz R. A., Skalka A. M., Leis J. The avian retroviral integration protein cleaves the terminal sequences of linear viral DNA at the in vivo sites of integration. J Virol. 1989 Dec;63(12):5319–5327. doi: 10.1128/jvi.63.12.5319-5327.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzman M., Mack J. P., Skalka A. M., Leis J. A covalent complex between retroviral integrase and nicked substrate DNA. Proc Natl Acad Sci U S A. 1991 Jun 1;88(11):4695–4699. doi: 10.1073/pnas.88.11.4695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan E., Mack J. P., Katz R. A., Kulkosky J., Skalka A. M. Retroviral integrase domains: DNA binding and the recognition of LTR sequences. Nucleic Acids Res. 1991 Feb 25;19(4):851–860. doi: 10.1093/nar/19.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkosky J., Jones K. S., Katz R. A., Mack J. P., Skalka A. M. Residues critical for retroviral integrative recombination in a region that is highly conserved among retroviral/retrotransposon integrases and bacterial insertion sequence transposases. Mol Cell Biol. 1992 May;12(5):2331–2338. doi: 10.1128/mcb.12.5.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaFemina R. L., Callahan P. L., Cordingley M. G. Substrate specificity of recombinant human immunodeficiency virus integrase protein. J Virol. 1991 Oct;65(10):5624–5630. doi: 10.1128/jvi.65.10.5624-5630.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaFemina R. L., Schneider C. L., Robbins H. L., Callahan P. L., LeGrow K., Roth E., Schleif W. A., Emini E. A. Requirement of active human immunodeficiency virus type 1 integrase enzyme for productive infection of human T-lymphoid cells. J Virol. 1992 Dec;66(12):7414–7419. doi: 10.1128/jvi.66.12.7414-7419.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leavitt A. D., Rose R. B., Varmus H. E. Both substrate and target oligonucleotide sequences affect in vitro integration mediated by human immunodeficiency virus type 1 integrase protein produced in Saccharomyces cerevisiae. J Virol. 1992 Apr;66(4):2359–2368. doi: 10.1128/jvi.66.4.2359-2368.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leavitt A. D., Shiue L., Varmus H. E. Site-directed mutagenesis of HIV-1 integrase demonstrates differential effects on integrase functions in vitro. J Biol Chem. 1993 Jan 25;268(3):2113–2119. [PubMed] [Google Scholar]
- Murphy J. E., Goff S. P. A mutation at one end of Moloney murine leukemia virus DNA blocks cleavage of both ends by the viral integrase in vivo. J Virol. 1992 Aug;66(8):5092–5095. doi: 10.1128/jvi.66.8.5092-5095.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathanson N., Georgsson G., Pálsson P. A., Najjar J. A., Lutley R., Pétursson G. Experimental visna in Icelandic sheep: the prototype lentiviral infection. Rev Infect Dis. 1985 Jan-Feb;7(1):75–82. doi: 10.1093/clinids/7.1.75. [DOI] [PubMed] [Google Scholar]
- Pahl A., Flügel R. M. Endonucleolytic cleavages and DNA-joining activities of the integration protein of human foamy virus. J Virol. 1993 Sep;67(9):5426–5434. doi: 10.1128/jvi.67.9.5426-5434.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pétursson G., Pálsson P. A., Georgsson G. Maedi-visna in sheep: host-virus interactions and utilization as a model. Intervirology. 1989;30 (Suppl 1):36–44. doi: 10.1159/000150122. [DOI] [PubMed] [Google Scholar]
- Schauer M., Billich A. The N-terminal region of HIV-1 integrase is required for integration activity, but not for DNA-binding. Biochem Biophys Res Commun. 1992 Jun 30;185(3):874–880. doi: 10.1016/0006-291x(92)91708-x. [DOI] [PubMed] [Google Scholar]
- Sherman P. A., Dickson M. L., Fyfe J. A. Human immunodeficiency virus type 1 integration protein: DNA sequence requirements for cleaving and joining reactions. J Virol. 1992 Jun;66(6):3593–3601. doi: 10.1128/jvi.66.6.3593-3601.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman P. A., Fyfe J. A. Human immunodeficiency virus integration protein expressed in Escherichia coli possesses selective DNA cleaving activity. Proc Natl Acad Sci U S A. 1990 Jul;87(13):5119–5123. doi: 10.1073/pnas.87.13.5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalka A. M. Retroviral proteases: first glimpses at the anatomy of a processing machine. Cell. 1989 Mar 24;56(6):911–913. doi: 10.1016/0092-8674(89)90621-1. [DOI] [PubMed] [Google Scholar]
- Sonigo P., Alizon M., Staskus K., Klatzmann D., Cole S., Danos O., Retzel E., Tiollais P., Haase A., Wain-Hobson S. Nucleotide sequence of the visna lentivirus: relationship to the AIDS virus. Cell. 1985 Aug;42(1):369–382. doi: 10.1016/s0092-8674(85)80132-x. [DOI] [PubMed] [Google Scholar]
- Staskus K. A., Retzel E. F., Lewis E. D., Silsby J. L., St Cyr S., Rank J. M., Wietgrefe S. W., Haase A. T., Cook R., Fast D. Isolation of replication-competent molecular clones of visna virus. Virology. 1991 Mar;181(1):228–240. doi: 10.1016/0042-6822(91)90488-w. [DOI] [PubMed] [Google Scholar]
- Vincent K. A., Ellison V., Chow S. A., Brown P. O. Characterization of human immunodeficiency virus type 1 integrase expressed in Escherichia coli and analysis of variants with amino-terminal mutations. J Virol. 1993 Jan;67(1):425–437. doi: 10.1128/jvi.67.1.425-437.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vink C., Oude Groeneger A. M., Plasterk R. H. Identification of the catalytic and DNA-binding region of the human immunodeficiency virus type I integrase protein. Nucleic Acids Res. 1993 Mar 25;21(6):1419–1425. doi: 10.1093/nar/21.6.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vink C., Yeheskiely E., van der Marel G. A., van Boom J. H., Plasterk R. H. Site-specific hydrolysis and alcoholysis of human immunodeficiency virus DNA termini mediated by the viral integrase protein. Nucleic Acids Res. 1991 Dec 25;19(24):6691–6698. doi: 10.1093/nar/19.24.6691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vink C., van Gent D. C., Elgersma Y., Plasterk R. H. Human immunodeficiency virus integrase protein requires a subterminal position of its viral DNA recognition sequence for efficient cleavage. J Virol. 1991 Sep;65(9):4636–4644. doi: 10.1128/jvi.65.9.4636-4644.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vora A. C., Fitzgerald M. L., Grandgenett D. P. Removal of 3'-OH-terminal nucleotides from blunt-ended long terminal repeat termini by the avian retrovirus integration protein. J Virol. 1990 Nov;64(11):5656–5659. doi: 10.1128/jvi.64.11.5656-5659.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woerner A. M., Klutch M., Levin J. G., Marcus-Sekura C. J. Localization of DNA binding activity of HIV-1 integrase to the C-terminal half of the protein. AIDS Res Hum Retroviruses. 1992 Feb;8(2):297–304. doi: 10.1089/aid.1992.8.297. [DOI] [PubMed] [Google Scholar]
- van Gent D. C., Elgersma Y., Bolk M. W., Vink C., Plasterk R. H. DNA binding properties of the integrase proteins of human immunodeficiency viruses types 1 and 2. Nucleic Acids Res. 1991 Jul 25;19(14):3821–3827. doi: 10.1093/nar/19.14.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gent D. C., Groeneger A. A., Plasterk R. H. Mutational analysis of the integrase protein of human immunodeficiency virus type 2. Proc Natl Acad Sci U S A. 1992 Oct 15;89(20):9598–9602. doi: 10.1073/pnas.89.20.9598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gent D. C., Vink C., Groeneger A. A., Plasterk R. H. Complementation between HIV integrase proteins mutated in different domains. EMBO J. 1993 Aug;12(8):3261–3267. doi: 10.1002/j.1460-2075.1993.tb05995.x. [DOI] [PMC free article] [PubMed] [Google Scholar]