Abstract
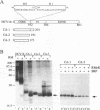
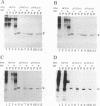
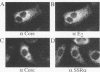
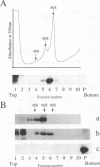
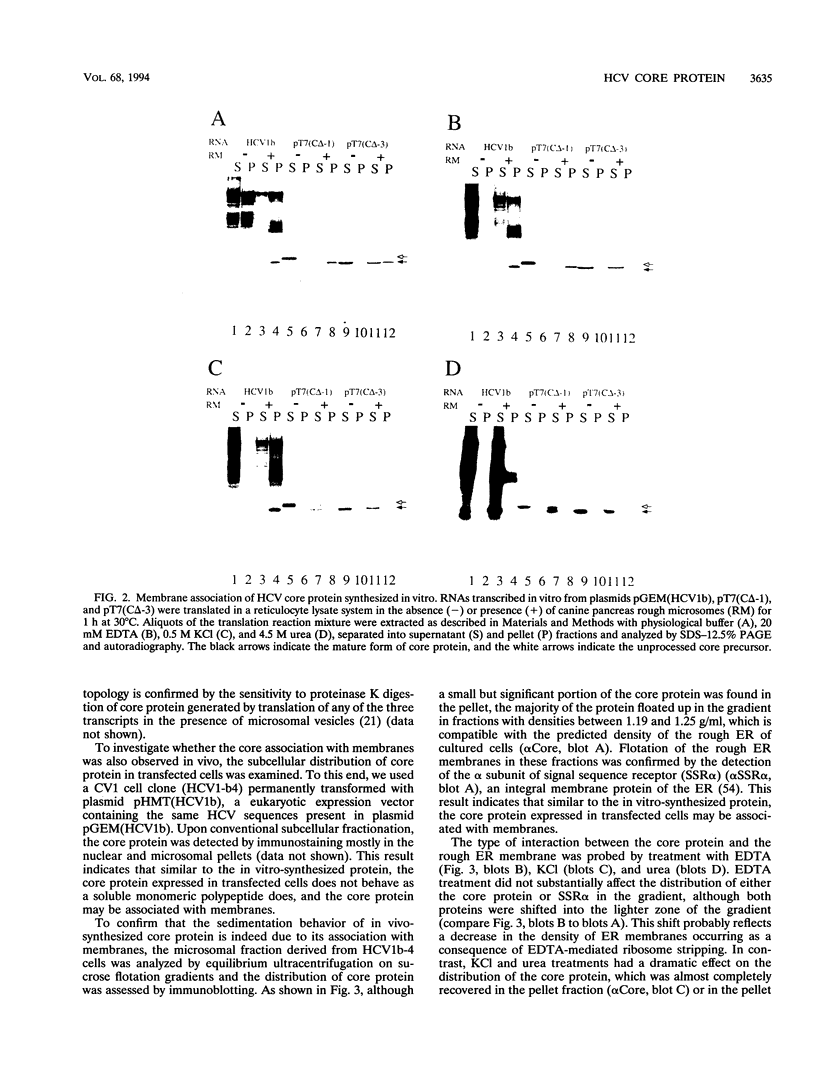
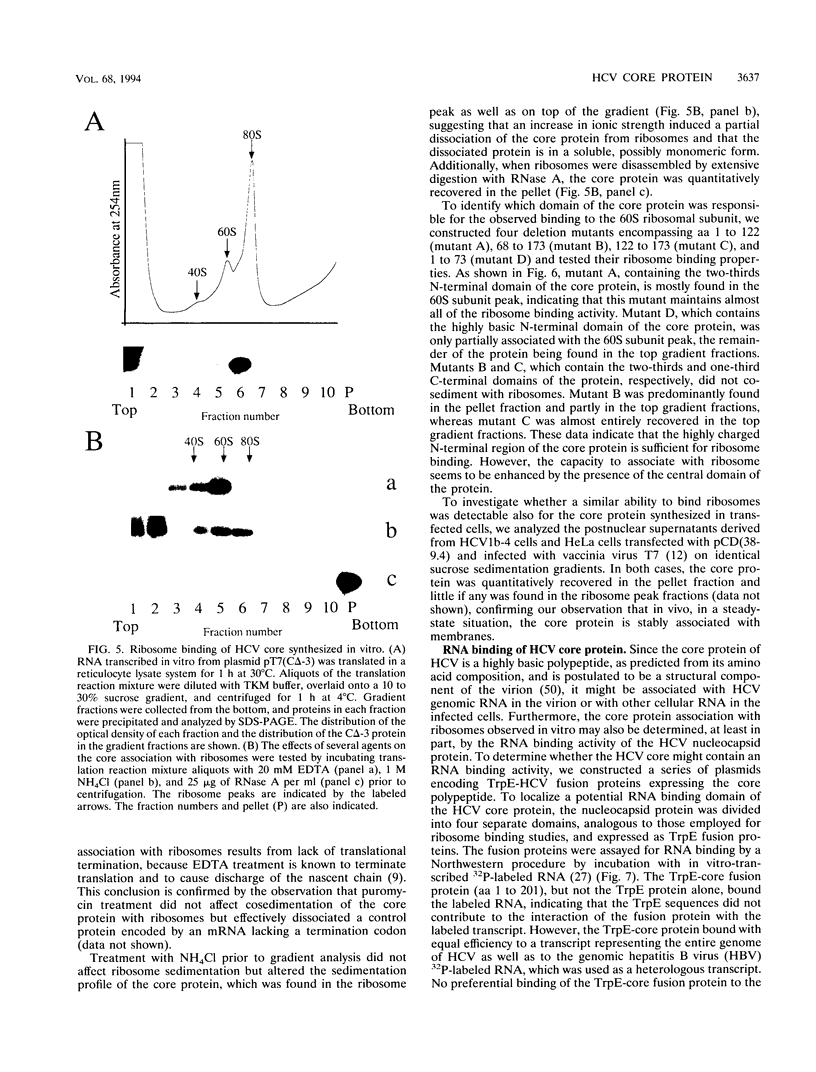
The biosynthesis and biochemical properties of the putative nucleocapsid protein of hepatitis C virus (HCV) were investigated. RNA transcripts for cell-free translation were prepared from truncated form of the cDNA construct encoding the structural proteins of HCV. Processing of the translation products was dependent on microsomal membranes and signal recognition particle, suggesting that release of the 21-kDa core protein from the polyprotein precursor is mediated solely by the signal peptidase of the endoplasmic reticulum (ER) and is achieved by the removal of a putative signal sequence of approximately 18 residues located at its C terminus. The core protein was found to bind membranes in vitro and in transfected cells, as shown by centrifugation analysis of in vitro translation products and transfected-cell lysates. Immunofluorescence of transfected cells showed that the core protein colocalized with the E2 glycoprotein as well as with a cellular ER membrane marker. The nucleocapsid protein expressed by in vitro translation in rabbit reticulocyte lysates cosedimented with the large ribosomal subunit in sucrose gradients. The ribosome binding domain was mapped to the N-terminal region of the core protein. Moreover, the same region was shown to bind RNA in vitro, suggesting that cosedimentation of core protein with ribosomes may be mediated by the RNA binding of the nucleocapsid protein of HCV. These studies indicate that the HCV core protein is a cytoplasmic protein associated with the ER membranes and possesses RNA binding activity.
Full text
PDF



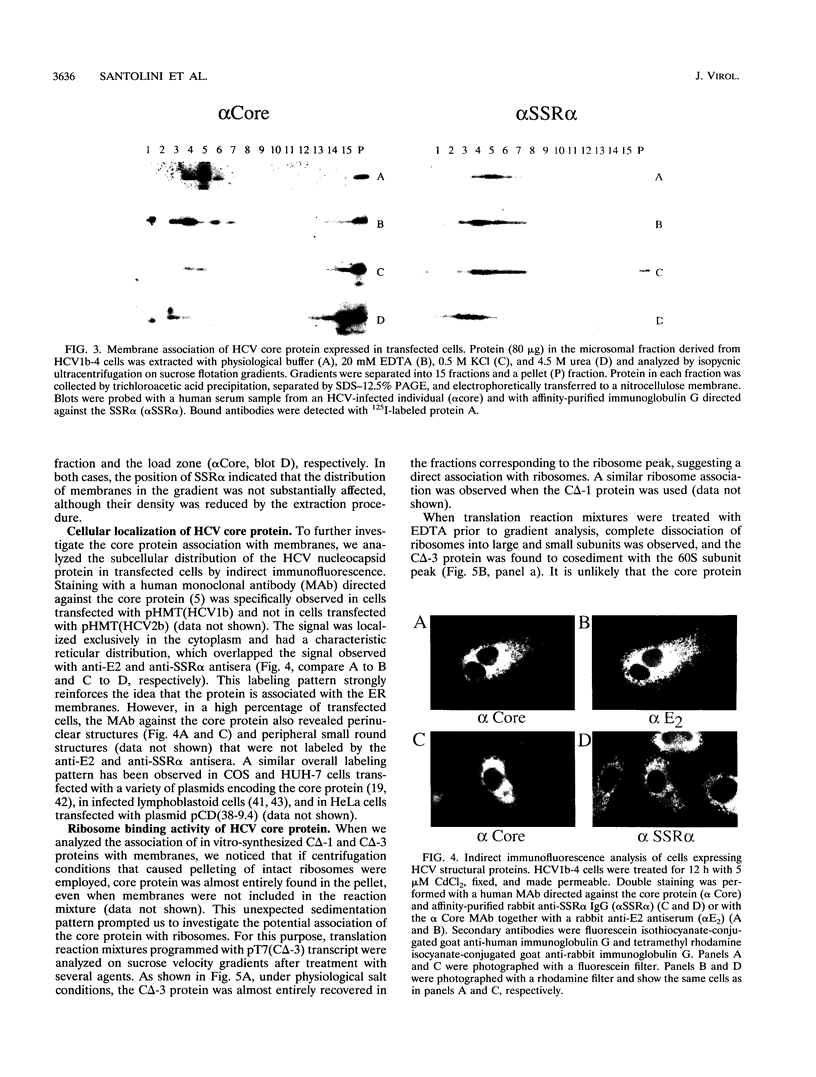




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abe K., Kurata T., Shikata T. Non-A, non-B hepatitis: visualization of virus-like particles from chimpanzee and human sera. Arch Virol. 1989;104(3-4):351–355. doi: 10.1007/BF01315557. [DOI] [PubMed] [Google Scholar]
- Bartenschlager R., Ahlborn-Laake L., Mous J., Jacobsen H. Nonstructural protein 3 of the hepatitis C virus encodes a serine-type proteinase required for cleavage at the NS3/4 and NS4/5 junctions. J Virol. 1993 Jul;67(7):3835–3844. doi: 10.1128/jvi.67.7.3835-3844.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartenschlager R., Junker-Niepmann M., Schaller H. The P gene product of hepatitis B virus is required as a structural component for genomic RNA encapsidation. J Virol. 1990 Nov;64(11):5324–5332. doi: 10.1128/jvi.64.11.5324-5332.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzeski H., Clegg J. C., Atkins G. J., Kennedy S. I. Regulation of the synthesis of Sindbis virus-specified RNA: role of the virion core protein. J Gen Virol. 1978 Mar;38(3):461–470. doi: 10.1099/0022-1317-38-3-461. [DOI] [PubMed] [Google Scholar]
- Cerino A., Boender P., La Monica N., Rosa C., Habets W., Mondelli M. U. A human monoclonal antibody specific for the N terminus of the hepatitis C virus nucleocapsid protein. J Immunol. 1993 Dec 15;151(12):7005–7015. [PubMed] [Google Scholar]
- Cha T. A., Beall E., Irvine B., Kolberg J., Chien D., Kuo G., Urdea M. S. At least five related, but distinct, hepatitis C viral genotypes exist. Proc Natl Acad Sci U S A. 1992 Aug 1;89(15):7144–7148. doi: 10.1073/pnas.89.15.7144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo Q. L., Kuo G., Weiner A. J., Overby L. R., Bradley D. W., Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989 Apr 21;244(4902):359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- Choo Q. L., Richman K. H., Han J. H., Berger K., Lee C., Dong C., Gallegos C., Coit D., Medina-Selby R., Barr P. J. Genetic organization and diversity of the hepatitis C virus. Proc Natl Acad Sci U S A. 1991 Mar 15;88(6):2451–2455. doi: 10.1073/pnas.88.6.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly T., Gilmore R. Formation of a functional ribosome-membrane junction during translocation requires the participation of a GTP-binding protein. J Cell Biol. 1986 Dec;103(6 Pt 1):2253–2261. doi: 10.1083/jcb.103.6.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley K. S., Reinhart G. D., Johnson A. E. The signal sequence moves through a ribosomal tunnel into a noncytoplasmic aqueous environment at the ER membrane early in translocation. Cell. 1993 Jun 18;73(6):1101–1115. doi: 10.1016/0092-8674(93)90640-c. [DOI] [PubMed] [Google Scholar]
- Fuerst T. R., Niles E. G., Studier F. W., Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci U S A. 1986 Nov;83(21):8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia P. D., Ou J. H., Rutter W. J., Walter P. Targeting of the hepatitis B virus precore protein to the endoplasmic reticulum membrane: after signal peptide cleavage translocation can be aborted and the product released into the cytoplasm. J Cell Biol. 1988 Apr;106(4):1093–1104. doi: 10.1083/jcb.106.4.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geigenmüller-Gnirke U., Nitschko H., Schlesinger S. Deletion analysis of the capsid protein of Sindbis virus: identification of the RNA binding region. J Virol. 1993 Mar;67(3):1620–1626. doi: 10.1128/jvi.67.3.1620-1626.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore R., Blobel G. Translocation of secretory proteins across the microsomal membrane occurs through an environment accessible to aqueous perturbants. Cell. 1985 Sep;42(2):497–505. doi: 10.1016/0092-8674(85)90107-2. [DOI] [PubMed] [Google Scholar]
- Gilmore R., Blobel G., Walter P. Protein translocation across the endoplasmic reticulum. I. Detection in the microsomal membrane of a receptor for the signal recognition particle. J Cell Biol. 1982 Nov;95(2 Pt 1):463–469. doi: 10.1083/jcb.95.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grakoui A., McCourt D. W., Wychowski C., Feinstone S. M., Rice C. M. Characterization of the hepatitis C virus-encoded serine proteinase: determination of proteinase-dependent polyprotein cleavage sites. J Virol. 1993 May;67(5):2832–2843. doi: 10.1128/jvi.67.5.2832-2843.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grakoui A., Wychowski C., Lin C., Feinstone S. M., Rice C. M. Expression and identification of hepatitis C virus polyprotein cleavage products. J Virol. 1993 Mar;67(3):1385–1395. doi: 10.1128/jvi.67.3.1385-1395.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada S., Watanabe Y., Takeuchi K., Suzuki T., Katayama T., Takebe Y., Saito I., Miyamura T. Expression of processed core protein of hepatitis C virus in mammalian cells. J Virol. 1991 Jun;65(6):3015–3021. doi: 10.1128/jvi.65.6.3015-3021.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatton T., Zhou S., Standring D. N. RNA- and DNA-binding activities in hepatitis B virus capsid protein: a model for their roles in viral replication. J Virol. 1992 Sep;66(9):5232–5241. doi: 10.1128/jvi.66.9.5232-5241.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hijikata M., Kato N., Ootsuyama Y., Nakagawa M., Shimotohno K. Gene mapping of the putative structural region of the hepatitis C virus genome by in vitro processing analysis. Proc Natl Acad Sci U S A. 1991 Jul 1;88(13):5547–5551. doi: 10.1073/pnas.88.13.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houghton M., Weiner A., Han J., Kuo G., Choo Q. L. Molecular biology of the hepatitis C viruses: implications for diagnosis, development and control of viral disease. Hepatology. 1991 Aug;14(2):381–388. [PubMed] [Google Scholar]
- Kato N., Hijikata M., Ootsuyama Y., Nakagawa M., Ohkoshi S., Sugimura T., Shimotohno K. Molecular cloning of the human hepatitis C virus genome from Japanese patients with non-A, non-B hepatitis. Proc Natl Acad Sci U S A. 1990 Dec;87(24):9524–9528. doi: 10.1073/pnas.87.24.9524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo G., Choo Q. L., Alter H. J., Gitnick G. L., Redeker A. G., Purcell R. H., Miyamura T., Dienstag J. L., Alter M. J., Stevens C. E. An assay for circulating antibodies to a major etiologic virus of human non-A, non-B hepatitis. Science. 1989 Apr 21;244(4902):362–364. doi: 10.1126/science.2496467. [DOI] [PubMed] [Google Scholar]
- Lee C. Z., Lin J. H., Chao M., McKnight K., Lai M. M. RNA-binding activity of hepatitis delta antigen involves two arginine-rich motifs and is required for hepatitis delta virus RNA replication. J Virol. 1993 Apr;67(4):2221–2227. doi: 10.1128/jvi.67.4.2221-2227.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljeström P., Garoff H. Internally located cleavable signal sequences direct the formation of Semliki Forest virus membrane proteins from a polyprotein precursor. J Virol. 1991 Jan;65(1):147–154. doi: 10.1128/jvi.65.1.147-154.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J. H., Chang M. F., Baker S. C., Govindarajan S., Lai M. M. Characterization of hepatitis delta antigen: specific binding to hepatitis delta virus RNA. J Virol. 1990 Sep;64(9):4051–4058. doi: 10.1128/jvi.64.9.4051-4058.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N., Brown D. T. Transient translocation of the cytoplasmic (endo) domain of a type I membrane glycoprotein into cellular membranes. J Cell Biol. 1993 Feb;120(4):877–883. doi: 10.1083/jcb.120.4.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobigs M. Flavivirus premembrane protein cleavage and spike heterodimer secretion require the function of the viral proteinase NS3. Proc Natl Acad Sci U S A. 1993 Jul 1;90(13):6218–6222. doi: 10.1073/pnas.90.13.6218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer D. I., Krause E., Dobberstein B. Secretory protein translocation across membranes-the role of the "docking protein'. Nature. 1982 Jun 24;297(5868):647–650. doi: 10.1038/297647a0. [DOI] [PubMed] [Google Scholar]
- Migliaccio G., Nicchitta C. V., Blobel G. The signal sequence receptor, unlike the signal recognition particle receptor, is not essential for protein translocation. J Cell Biol. 1992 Apr;117(1):15–25. doi: 10.1083/jcb.117.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller R. H., Purcell R. H. Hepatitis C virus shares amino acid sequence similarity with pestiviruses and flaviviruses as well as members of two plant virus supergroups. Proc Natl Acad Sci U S A. 1990 Mar;87(6):2057–2061. doi: 10.1073/pnas.87.6.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicchitta C. V., Blobel G. Lumenal proteins of the mammalian endoplasmic reticulum are required to complete protein translocation. Cell. 1993 Jun 4;73(5):989–998. doi: 10.1016/0092-8674(93)90276-v. [DOI] [PubMed] [Google Scholar]
- Nowak T., Färber P. M., Wengler G., Wengler G. Analyses of the terminal sequences of West Nile virus structural proteins and of the in vitro translation of these proteins allow the proposal of a complete scheme of the proteolytic cleavages involved in their synthesis. Virology. 1989 Apr;169(2):365–376. doi: 10.1016/0042-6822(89)90162-1. [DOI] [PubMed] [Google Scholar]
- Ooi C. E., Weiss J. Bidirectional movement of a nascent polypeptide across microsomal membranes reveals requirements for vectorial translocation of proteins. Cell. 1992 Oct 2;71(1):87–96. doi: 10.1016/0092-8674(92)90268-h. [DOI] [PubMed] [Google Scholar]
- Pizzi E., Tramontano A., Tomei L., La Monica N., Failla C., Sardana M., Wood T., De Francesco R. Molecular model of the specificity pocket of the hepatitis C virus protease: implications for substrate recognition. Proc Natl Acad Sci U S A. 1994 Feb 1;91(3):888–892. doi: 10.1073/pnas.91.3.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapoport T. A. Transport of proteins across the endoplasmic reticulum membrane. Science. 1992 Nov 6;258(5084):931–936. doi: 10.1126/science.1332192. [DOI] [PubMed] [Google Scholar]
- Rümenapf T., Unger G., Strauss J. H., Thiel H. J. Processing of the envelope glycoproteins of pestiviruses. J Virol. 1993 Jun;67(6):3288–3294. doi: 10.1128/jvi.67.6.3288-3294.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini D. D., Tashiro Y., Palade G. E. On the attachment of ribosomes to microsomal membranes. J Mol Biol. 1966 Aug;19(2):503–524. doi: 10.1016/s0022-2836(66)80019-0. [DOI] [PubMed] [Google Scholar]
- Schmidt C. J., Jubier M. F., Hamer D. H. Structure and expression of two human metallothionein-I isoform genes and a related pseudogene. J Biol Chem. 1985 Jun 25;260(12):7731–7737. [PubMed] [Google Scholar]
- Selby M. J., Choo Q. L., Berger K., Kuo G., Glazer E., Eckart M., Lee C., Chien D., Kuo C., Houghton M. Expression, identification and subcellular localization of the proteins encoded by the hepatitis C viral genome. J Gen Virol. 1993 Jun;74(Pt 6):1103–1113. doi: 10.1099/0022-1317-74-6-1103. [DOI] [PubMed] [Google Scholar]
- Shih C. M., Lo S. J., Miyamura T., Chen S. Y., Lee Y. H. Suppression of hepatitis B virus expression and replication by hepatitis C virus core protein in HuH-7 cells. J Virol. 1993 Oct;67(10):5823–5832. doi: 10.1128/jvi.67.10.5823-5832.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu Y. K., Iwamoto A., Hijikata M., Purcell R. H., Yoshikura H. Evidence for in vitro replication of hepatitis C virus genome in a human T-cell line. Proc Natl Acad Sci U S A. 1992 Jun 15;89(12):5477–5481. doi: 10.1073/pnas.89.12.5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon S. M., Peskin C. S., Oster G. F. What drives the translocation of proteins? Proc Natl Acad Sci U S A. 1992 May 1;89(9):3770–3774. doi: 10.1073/pnas.89.9.3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh I., Helenius A. Role of ribosomes in Semliki Forest virus nucleocapsid uncoating. J Virol. 1992 Dec;66(12):7049–7058. doi: 10.1128/jvi.66.12.7049-7058.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spindler K. R., Rosser D. S., Berk A. J. Analysis of adenovirus transforming proteins from early regions 1A and 1B with antisera to inducible fusion antigens produced in Escherichia coli. J Virol. 1984 Jan;49(1):132–141. doi: 10.1128/jvi.49.1.132-141.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark R., Meyers G., Rümenapf T., Thiel H. J. Processing of pestivirus polyprotein: cleavage site between autoprotease and nucleocapsid protein of classical swine fever virus. J Virol. 1993 Dec;67(12):7088–7095. doi: 10.1128/jvi.67.12.7088-7095.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamizawa A., Mori C., Fuke I., Manabe S., Murakami S., Fujita J., Onishi E., Andoh T., Yoshida I., Okayama H. Structure and organization of the hepatitis C virus genome isolated from human carriers. J Virol. 1991 Mar;65(3):1105–1113. doi: 10.1128/jvi.65.3.1105-1113.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi K., Kubo Y., Boonmar S., Watanabe Y., Katayama T., Choo Q. L., Kuo G., Houghton M., Saito I., Miyamura T. The putative nucleocapsid and envelope protein genes of hepatitis C virus determined by comparison of the nucleotide sequences of two isolates derived from an experimentally infected chimpanzee and healthy human carriers. J Gen Virol. 1990 Dec;71(Pt 12):3027–3033. doi: 10.1099/0022-1317-71-12-3027. [DOI] [PubMed] [Google Scholar]
- Tomei L., Failla C., Santolini E., De Francesco R., La Monica N. NS3 is a serine protease required for processing of hepatitis C virus polyprotein. J Virol. 1993 Jul;67(7):4017–4026. doi: 10.1128/jvi.67.7.4017-4026.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmanen I., Söderlund H., Käriäinen L. Role of protein synthesis in the assembly of Semliki forest virus nucleocapsid. Virology. 1979 Dec;99(2):265–276. doi: 10.1016/0042-6822(79)90006-0. [DOI] [PubMed] [Google Scholar]
- Vogel F., Hartmann E., Görlich D., Rapoport T. A. Segregation of the signal sequence receptor protein in the rough endoplasmic reticulum membrane. Eur J Cell Biol. 1990 Dec;53(2):197–202. [PubMed] [Google Scholar]
- Walter P., Blobel G. Translocation of proteins across the endoplasmic reticulum. II. Signal recognition protein (SRP) mediates the selective binding to microsomal membranes of in-vitro-assembled polysomes synthesizing secretory protein. J Cell Biol. 1981 Nov;91(2 Pt 1):551–556. doi: 10.1083/jcb.91.2.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P., Ibrahimi I., Blobel G. Translocation of proteins across the endoplasmic reticulum. I. Signal recognition protein (SRP) binds to in-vitro-assembled polysomes synthesizing secretory protein. J Cell Biol. 1981 Nov;91(2 Pt 1):545–550. doi: 10.1083/jcb.91.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P., Lingappa V. R. Mechanism of protein translocation across the endoplasmic reticulum membrane. Annu Rev Cell Biol. 1986;2:499–516. doi: 10.1146/annurev.cb.02.110186.002435. [DOI] [PubMed] [Google Scholar]
- Weeks K. M., Ampe C., Schultz S. C., Steitz T. A., Crothers D. M. Fragments of the HIV-1 Tat protein specifically bind TAR RNA. Science. 1990 Sep 14;249(4974):1281–1285. doi: 10.1126/science.2205002. [DOI] [PubMed] [Google Scholar]
- Wengler G. The mode of assembly of alphavirus cores implies a mechanism for the disassembly of the cores in the early stages of infection. Brief review. Arch Virol. 1987;94(1-2):1–14. doi: 10.1007/BF01313721. [DOI] [PubMed] [Google Scholar]
- Wengler G., Wengler G., Boege U., Wahn K. Establishment and analysis of a system which allows assembly and disassembly of alphavirus core-like particles under physiological conditions in vitro. Virology. 1984 Jan 30;132(2):401–412. doi: 10.1016/0042-6822(84)90045-x. [DOI] [PubMed] [Google Scholar]
- Wengler G., Wengler G. Identification of a transfer of viral core protein to cellular ribosomes during the early stages of alphavirus infection. Virology. 1984 Apr 30;134(2):435–442. doi: 10.1016/0042-6822(84)90310-6. [DOI] [PubMed] [Google Scholar]
- Wengler G., Würkner D., Wengler G. Identification of a sequence element in the alphavirus core protein which mediates interaction of cores with ribosomes and the disassembly of cores. Virology. 1992 Dec;191(2):880–888. doi: 10.1016/0042-6822(92)90263-o. [DOI] [PubMed] [Google Scholar]
- van Steeg H., Kasperaitis M., Voorma H. O., Benne R. Infection of neuroblastoma cells by Semliki Forest virus. The interference of viral capsid protein with the binding of host messenger RNAs into initiation complexes is the cause of the shut-off of host protein synthesis. Eur J Biochem. 1984 Feb 1;138(3):473–478. doi: 10.1111/j.1432-1033.1984.tb07940.x. [DOI] [PubMed] [Google Scholar]