Abstract
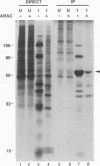
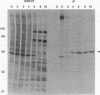
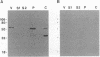
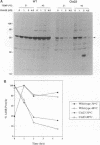
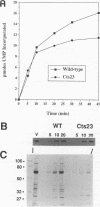
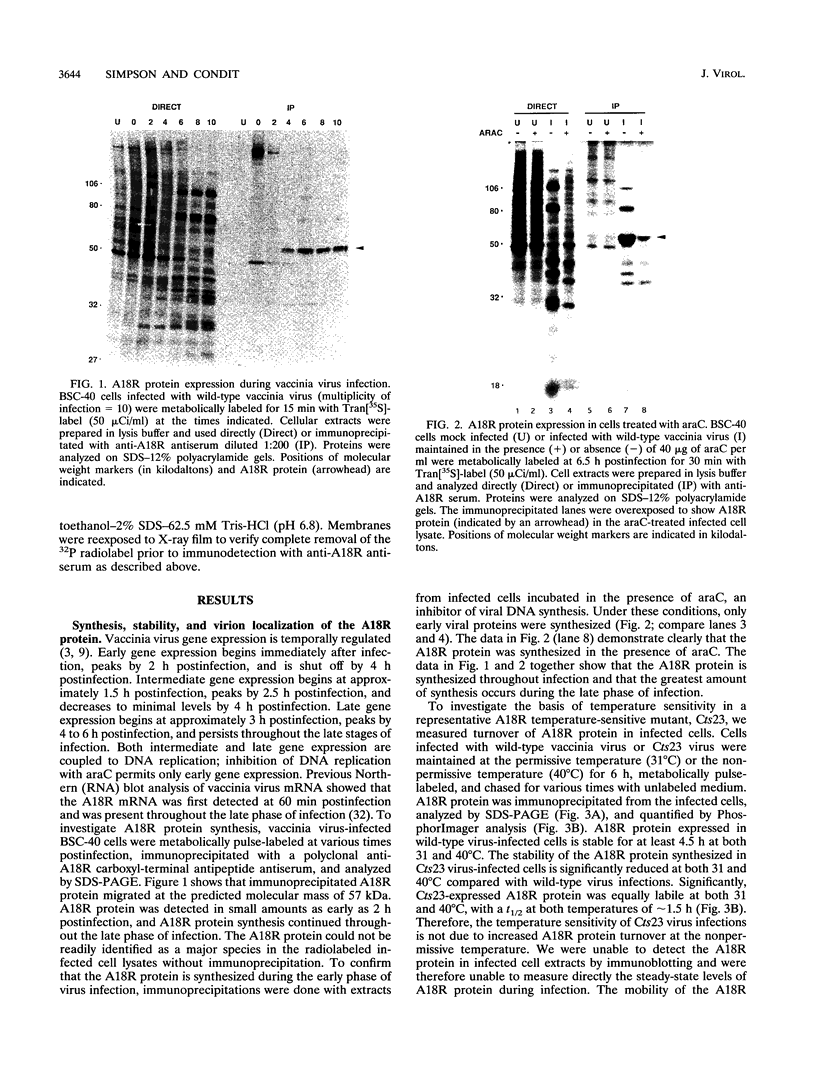
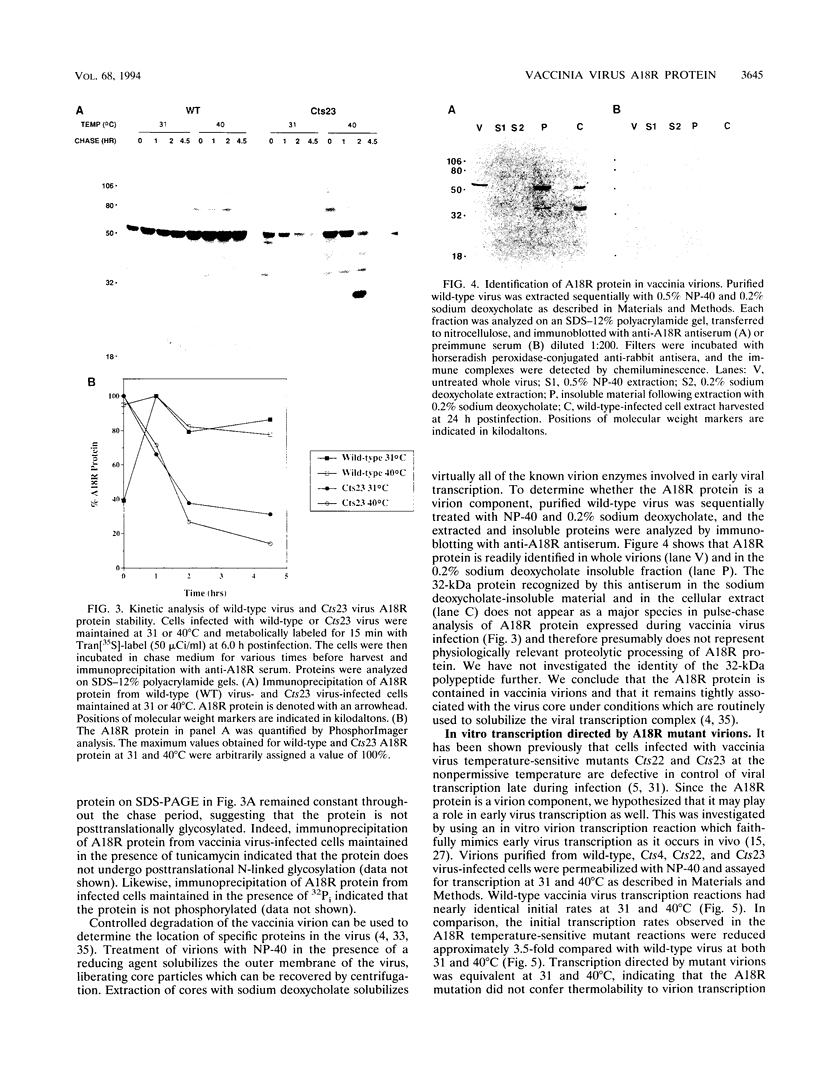
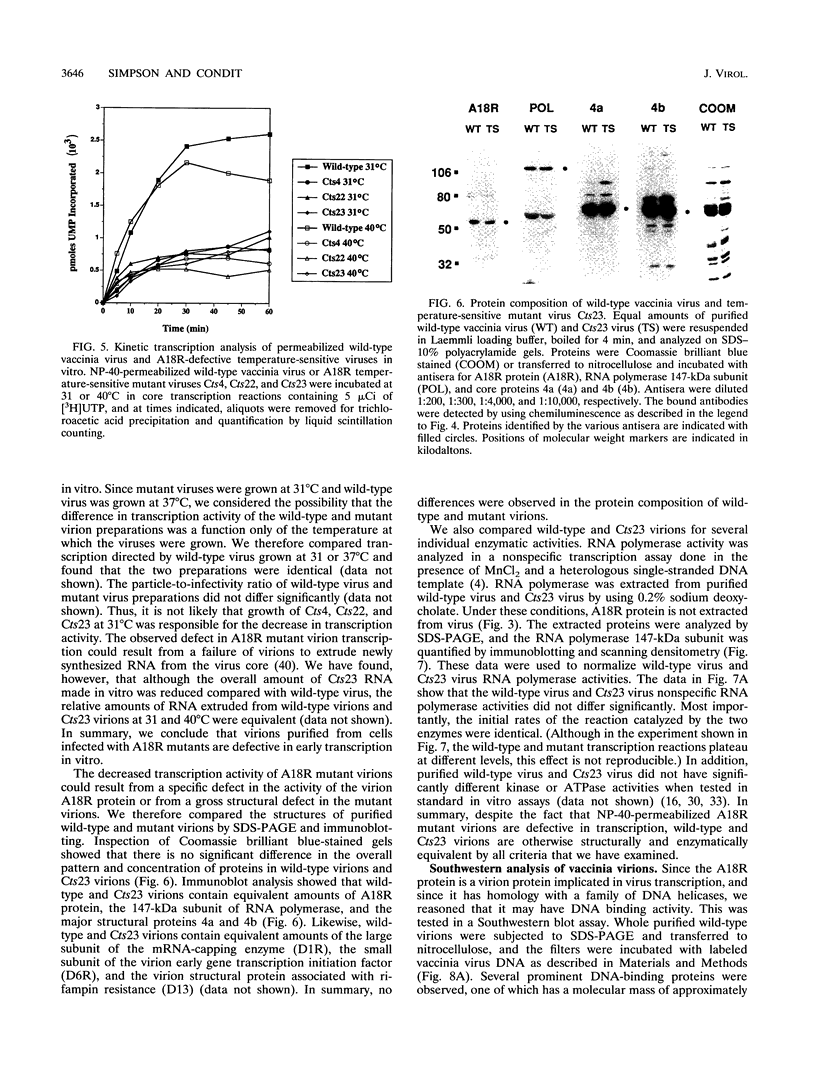
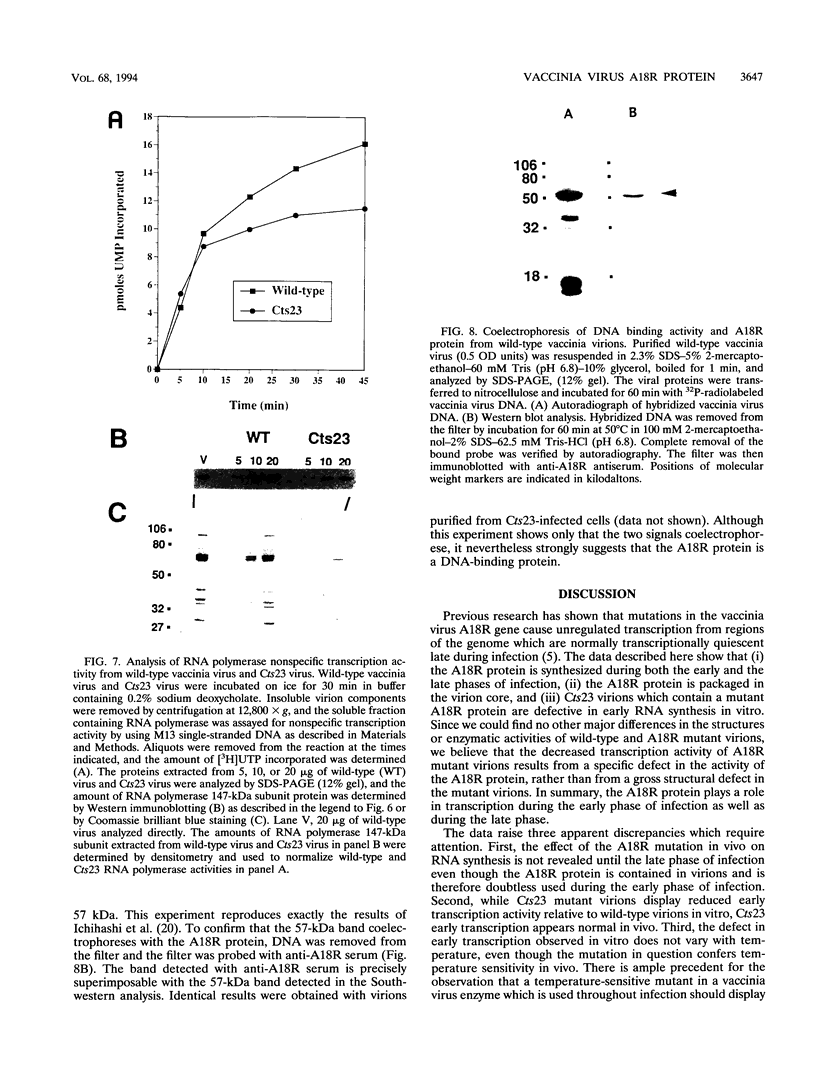
The vaccinia virus gene A18R is essential for virus infection. The loss of A18R protein function results in unregulated transcription late during virus infection from regions of the viral genome which are normally transcriptionally quiescent. We have characterized A18R protein expression in cells infected with wild-type virus and the A18R temperature-sensitive mutant Cts23. The A18R protein is expressed during early and late phases of infection. The A18R protein expressed by Cts23 virus at permissive and nonpermissive temperatures is significantly less stable than the wild-type A18R protein. The A18R protein was identified as a virion component and localized by detergent extraction to the virion core. Virions purified from cells infected with the A18R temperature-sensitive mutants Cts4, Cts22, and Cts23 are defective in early viral transcription in vitro. The mutant transcription defect is not attributable to gross defects in virion structure or virion DNA-dependent RNA polymerase activity. We conclude that the A18R protein plays a role in viral transcription during the early phase of infection as well as during the late phase.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ahn B. Y., Moss B. RNA polymerase-associated transcription specificity factor encoded by vaccinia virus. Proc Natl Acad Sci U S A. 1992 Apr 15;89(8):3536–3540. doi: 10.1073/pnas.89.8.3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldick C. J., Jr, Keck J. G., Moss B. Mutational analysis of the core, spacer, and initiator regions of vaccinia virus intermediate-class promoters. J Virol. 1992 Aug;66(8):4710–4719. doi: 10.1128/jvi.66.8.4710-4719.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldick C. J., Jr, Moss B. Characterization and temporal regulation of mRNAs encoded by vaccinia virus intermediate-stage genes. J Virol. 1993 Jun;67(6):3515–3527. doi: 10.1128/jvi.67.6.3515-3527.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baroudy B. M., Moss B. Purification and characterization of a DNA-dependent RNA polymerase from vaccinia virions. J Biol Chem. 1980 May 10;255(9):4372–4380. [PubMed] [Google Scholar]
- Bayliss C. D., Condit R. C. Temperature-sensitive mutants in the vaccinia virus A18R gene increase double-stranded RNA synthesis as a result of aberrant viral transcription. Virology. 1993 May;194(1):254–262. doi: 10.1006/viro.1993.1256. [DOI] [PubMed] [Google Scholar]
- Broyles S. S., Yuen L., Shuman S., Moss B. Purification of a factor required for transcription of vaccinia virus early genes. J Biol Chem. 1988 Aug 5;263(22):10754–10760. [PubMed] [Google Scholar]
- Buratowski S. DNA repair and transcription: the helicase connection. Science. 1993 Apr 2;260(5104):37–38. doi: 10.1126/science.8465198. [DOI] [PubMed] [Google Scholar]
- Condit R. C., Motyczka A. Isolation and preliminary characterization of temperature-sensitive mutants of vaccinia virus. Virology. 1981 Aug;113(1):224–241. doi: 10.1016/0042-6822(81)90150-1. [DOI] [PubMed] [Google Scholar]
- Condit R. C., Motyczka A., Spizz G. Isolation, characterization, and physical mapping of temperature-sensitive mutants of vaccinia virus. Virology. 1983 Jul 30;128(2):429–443. doi: 10.1016/0042-6822(83)90268-4. [DOI] [PubMed] [Google Scholar]
- Condit R. C., Niles E. G. Orthopoxvirus genetics. Curr Top Microbiol Immunol. 1990;163:1–39. doi: 10.1007/978-3-642-75605-4_1. [DOI] [PubMed] [Google Scholar]
- Davison A. J., Moss B. Structure of vaccinia virus early promoters. J Mol Biol. 1989 Dec 20;210(4):749–769. doi: 10.1016/0022-2836(89)90107-1. [DOI] [PubMed] [Google Scholar]
- Davison A. J., Moss B. Structure of vaccinia virus late promoters. J Mol Biol. 1989 Dec 20;210(4):771–784. doi: 10.1016/0022-2836(89)90108-3. [DOI] [PubMed] [Google Scholar]
- Dieckmann C. L., Tzagoloff A. Assembly of the mitochondrial membrane system. CBP6, a yeast nuclear gene necessary for synthesis of cytochrome b. J Biol Chem. 1985 Feb 10;260(3):1513–1520. [PubMed] [Google Scholar]
- Friedberg E. C. Xeroderma pigmentosum, Cockayne's syndrome, helicases, and DNA repair: what's the relationship? Cell. 1992 Dec 11;71(6):887–889. doi: 10.1016/0092-8674(92)90384-o. [DOI] [PubMed] [Google Scholar]
- Gershowitz A., Moss B. Abortive transcription products of vaccinia virus are guanylylated, methylated, and polyadenylylated. J Virol. 1979 Sep;31(3):849–853. doi: 10.1128/jvi.31.3.849-853.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold P. H., Dales S. Localization of nucleotide phosphohydrolase activity within vaccinia. Proc Natl Acad Sci U S A. 1968 Jul;60(3):845–852. doi: 10.1073/pnas.60.3.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooda-Dhingra U., Patel D. D., Pickup D. J., Condit R. C. Fine structure mapping and phenotypic analysis of five temperature-sensitive mutations in the second largest subunit of vaccinia virus DNA-dependent RNA polymerase. Virology. 1990 Jan;174(1):60–69. doi: 10.1016/0042-6822(90)90054-u. [DOI] [PubMed] [Google Scholar]
- Hooda-Dhingra U., Thompson C. L., Condit R. C. Detailed phenotypic characterization of five temperature-sensitive mutants in the 22- and 147-kilodalton subunits of vaccinia virus DNA-dependent RNA polymerase. J Virol. 1989 Feb;63(2):714–729. doi: 10.1128/jvi.63.2.714-729.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichihashi Y., Oie M., Tsuruhara T. Location of DNA-binding proteins and disulfide-linked proteins in vaccinia virus structural elements. J Virol. 1984 Jun;50(3):929–938. doi: 10.1128/jvi.50.3.929-938.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JOKLIK W. K. The purification fo four strains of poxvirus. Virology. 1962 Sep;18:9–18. doi: 10.1016/0042-6822(62)90172-1. [DOI] [PubMed] [Google Scholar]
- Keck J. G., Baldick C. J., Jr, Moss B. Role of DNA replication in vaccinia virus gene expression: a naked template is required for transcription of three late trans-activator genes. Cell. 1990 Jun 1;61(5):801–809. doi: 10.1016/0092-8674(90)90190-p. [DOI] [PubMed] [Google Scholar]
- Keck J. G., Kovacs G. R., Moss B. Overexpression, purification, and late transcription factor activity of the 17-kilodalton protein encoded by the vaccinia virus A1L gene. J Virol. 1993 Oct;67(10):5740–5748. doi: 10.1128/jvi.67.10.5740-5748.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E. V., Senkevich T. G. Vaccinia virus encodes four putative DNA and/or RNA helicases distantly related to each other. J Gen Virol. 1992 Apr;73(Pt 4):989–993. doi: 10.1099/0022-1317-73-4-989. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Moss B., Ahn B. Y., Amegadzie B., Gershon P. D., Keck J. G. Cytoplasmic transcription system encoded by vaccinia virus. J Biol Chem. 1991 Jan 25;266(3):1355–1358. [PubMed] [Google Scholar]
- Moss B. Regulation of vaccinia virus transcription. Annu Rev Biochem. 1990;59:661–688. doi: 10.1146/annurev.bi.59.070190.003305. [DOI] [PubMed] [Google Scholar]
- Moyer R. W. The role of the host cell nucleus in vaccinia virus morphogenesis. Virus Res. 1987 Sep;8(3):173–191. doi: 10.1016/0168-1702(87)90014-1. [DOI] [PubMed] [Google Scholar]
- Munyon W., Paoletti E., Ospina J., Grace J. T., Jr Nucleotide phosphohydrolase in purified vaccinia virus. J Virol. 1968 Mar;2(3):167–172. doi: 10.1128/jvi.2.3.167-172.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacha R. F., Condit R. C. Characterization of a temperature-sensitive mutant of vaccinia virus reveals a novel function that prevents virus-induced breakdown of RNA. J Virol. 1985 Nov;56(2):395–403. doi: 10.1128/jvi.56.2.395-403.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacha R. F., Meis R. J., Condit R. C. Structure and expression of the vaccinia virus gene which prevents virus-induced breakdown of RNA. J Virol. 1990 Aug;64(8):3853–3863. doi: 10.1128/jvi.64.8.3853-3863.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paoletti E., Moss B. Protein kinase and specific phosphate acceptor proteins associated with vaccinia virus cores. J Virol. 1972 Sep;10(3):417–424. doi: 10.1128/jvi.10.3.417-424.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvin J. D., Sharp P. A. DNA topology and a minimal set of basal factors for transcription by RNA polymerase II. Cell. 1993 May 7;73(3):533–540. doi: 10.1016/0092-8674(93)90140-l. [DOI] [PubMed] [Google Scholar]
- Rohrmann G., Moss B. Transcription of vaccinia virus early genes by a template-dependent soluble extract of purified virions. J Virol. 1985 Nov;56(2):349–355. doi: 10.1128/jvi.56.2.349-355.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SADLER J. R., NOVICK A. THE PROPERTIES OF REPRESSOR AND THE KINETICS OF ITS ACTION. J Mol Biol. 1965 Jun;12:305–327. doi: 10.1016/s0022-2836(65)80255-8. [DOI] [PubMed] [Google Scholar]
- Schaeffer L., Roy R., Humbert S., Moncollin V., Vermeulen W., Hoeijmakers J. H., Chambon P., Egly J. M. DNA repair helicase: a component of BTF2 (TFIIH) basic transcription factor. Science. 1993 Apr 2;260(5104):58–63. doi: 10.1126/science.8465201. [DOI] [PubMed] [Google Scholar]
- Selby C. P., Sancar A. Molecular mechanism of transcription-repair coupling. Science. 1993 Apr 2;260(5104):53–58. doi: 10.1126/science.8465200. [DOI] [PubMed] [Google Scholar]
- Shuman S., Moss B. Bromouridine triphosphate inhibits transcription termination and mRNA release by vaccinia virions. J Biol Chem. 1989 Dec 15;264(35):21356–21360. [PubMed] [Google Scholar]
- Thompson C. L., Hooda-Dhingra U., Condit R. C. Fine structure mapping of five temperature-sensitive mutants in the 22- and 147-kilodalton subunits of vaccinia virus DNA-dependent RNA polymerase. J Virol. 1989 Feb;63(2):705–713. doi: 10.1128/jvi.63.2.705-713.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos J. C., Sasker M., Stunnenberg H. G. Vaccinia virus capping enzyme is a transcription initiation factor. EMBO J. 1991 Sep;10(9):2553–2558. doi: 10.1002/j.1460-2075.1991.tb07795.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M. H., King J. Surface amino acids as sites of temperature-sensitive folding mutations in the P22 tailspike protein. J Biol Chem. 1988 Jan 25;263(3):1424–1431. [PubMed] [Google Scholar]
- Yuwen H., Cox J. H., Yewdell J. W., Bennink J. R., Moss B. Nuclear localization of a double-stranded RNA-binding protein encoded by the vaccinia virus E3L gene. Virology. 1993 Aug;195(2):732–744. doi: 10.1006/viro.1993.1424. [DOI] [PubMed] [Google Scholar]