Abstract
We have used a full-length cDNA clone of a mouse hepatitis virus strain A59 defective interfering (DI) RNA, pMIDI-C, and cassette mutagenesis to study the mechanism of coronavirus subgenomic mRNA synthesis. Promoter sequences closely resembling those of subgenomic mRNAs 3 and 7 were inserted into MIDI-C. Both subgenomic RNA promoters gave rise to the synthesis of a subgenomic DI RNA in virus-infected and DI RNA-transfected cells. From a mutagenic analysis of the promoters we concluded the following. (i) The extent of base pairing between the leader RNA and the intergenic promoter sequence does not control subgenomic RNA abundance. (ii) Promoter recognition does not rely on base pairing only. Presumably, transcription initiation requires recognition of the promoter sequence by the transcriptase. (iii) Fusion of leader and body sequences takes place at multiple--possibly random--sites within the intergenic promoter sequence. A model is presented in which, prior to elongation, the leader RNA is trimmed by a processive 3'-->5' nuclease.
Full text
PDF

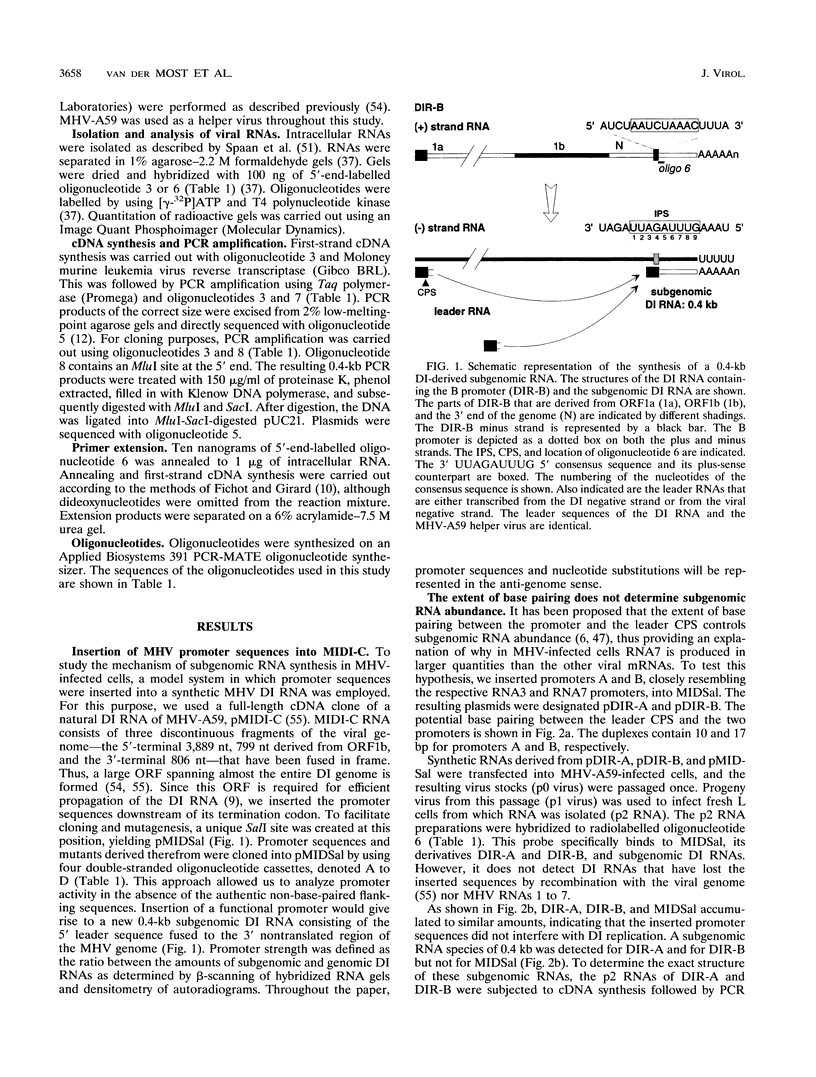


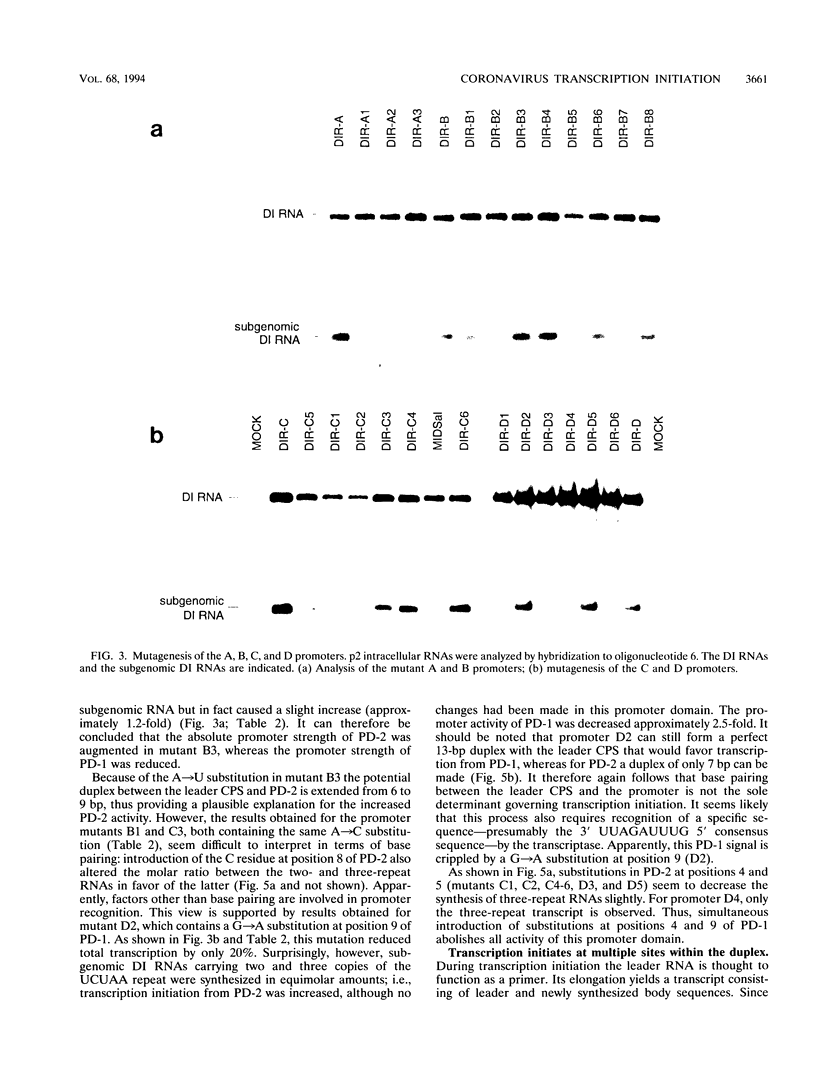
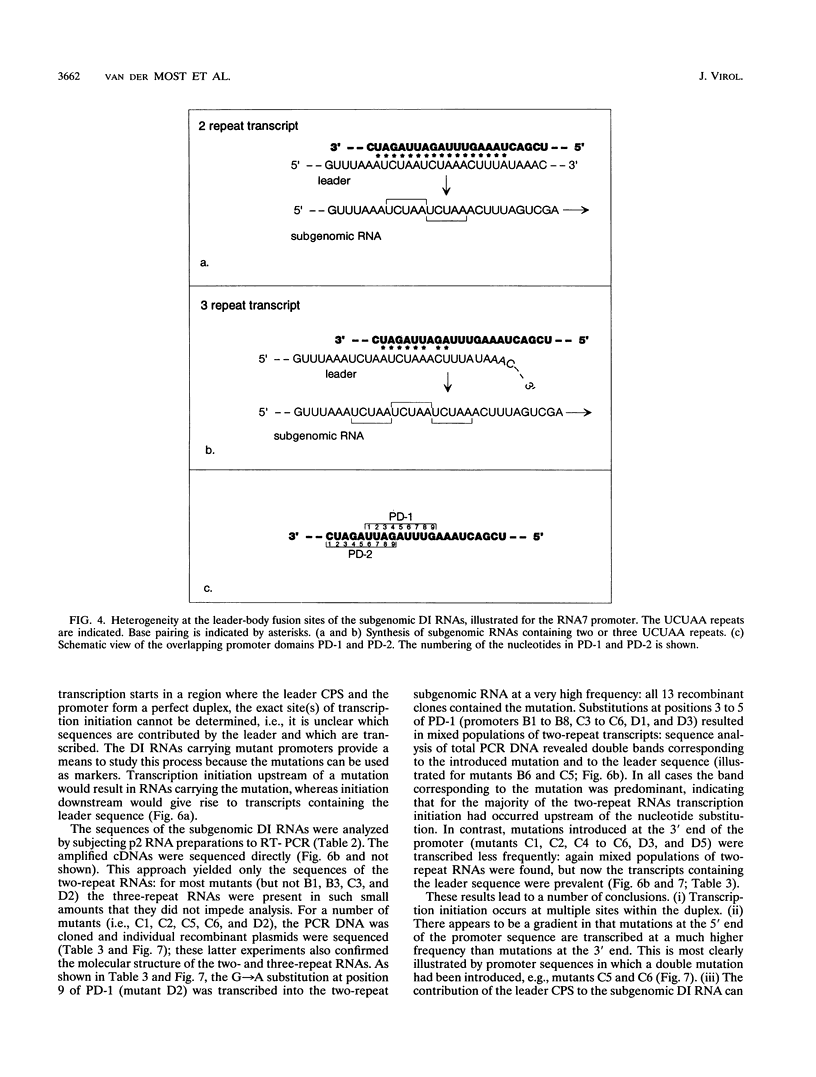
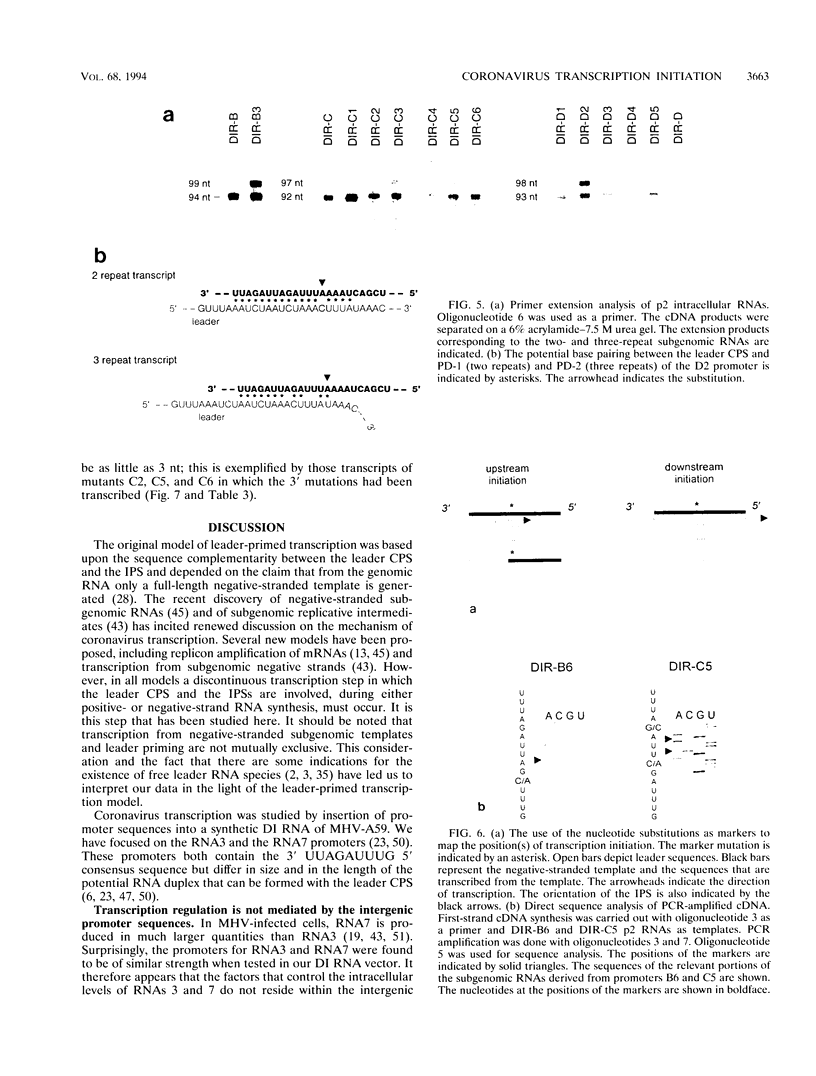


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Armstrong J., Niemann H., Smeekens S., Rottier P., Warren G. Sequence and topology of a model intracellular membrane protein, E1 glycoprotein, from a coronavirus. Nature. 1984 Apr 19;308(5961):751–752. doi: 10.1038/308751a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker S. C., Lai M. M. An in vitro system for the leader-primed transcription of coronavirus mRNAs. EMBO J. 1990 Dec;9(12):4173–4179. doi: 10.1002/j.1460-2075.1990.tb07641.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baric R. S., Stohlman S. A., Razavi M. K., Lai M. M. Characterization of leader-related small RNAs in coronavirus-infected cells: further evidence for leader-primed mechanism of transcription. Virus Res. 1985 Jul;3(1):19–33. doi: 10.1016/0168-1702(85)90038-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boursnell M. E., Brown T. D., Foulds I. J., Green P. F., Tomley F. M., Binns M. M. Completion of the sequence of the genome of the coronavirus avian infectious bronchitis virus. J Gen Virol. 1987 Jan;68(Pt 1):57–77. doi: 10.1099/0022-1317-68-1-57. [DOI] [PubMed] [Google Scholar]
- Bredenbeek P. J., Pachuk C. J., Noten A. F., Charité J., Luytjes W., Weiss S. R., Spaan W. J. The primary structure and expression of the second open reading frame of the polymerase gene of the coronavirus MHV-A59; a highly conserved polymerase is expressed by an efficient ribosomal frameshifting mechanism. Nucleic Acids Res. 1990 Apr 11;18(7):1825–1832. doi: 10.1093/nar/18.7.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budzilowicz C. J., Wilczynski S. P., Weiss S. R. Three intergenic regions of coronavirus mouse hepatitis virus strain A59 genome RNA contain a common nucleotide sequence that is homologous to the 3' end of the viral mRNA leader sequence. J Virol. 1985 Mar;53(3):834–840. doi: 10.1128/jvi.53.3.834-840.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fichot O., Girard M. An improved method for sequencing of RNA templates. Nucleic Acids Res. 1990 Oct 25;18(20):6162–6162. doi: 10.1093/nar/18.20.6162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French R., Ahlquist P. Characterization and engineering of sequences controlling in vivo synthesis of brome mosaic virus subgenomic RNA. J Virol. 1988 Jul;62(7):2411–2420. doi: 10.1128/jvi.62.7.2411-2420.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann M. A., Sethna P. B., Brian D. A. Bovine coronavirus mRNA replication continues throughout persistent infection in cell culture. J Virol. 1990 Sep;64(9):4108–4114. doi: 10.1128/jvi.64.9.4108-4114.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holbrook S. R., Cheong C., Tinoco I., Jr, Kim S. H. Crystal structure of an RNA double helix incorporating a track of non-Watson-Crick base pairs. Nature. 1991 Oct 10;353(6344):579–581. doi: 10.1038/353579a0. [DOI] [PubMed] [Google Scholar]
- Huang A. S., Baltimore D. Defective viral particles and viral disease processes. Nature. 1970 Apr 25;226(5243):325–327. doi: 10.1038/226325a0. [DOI] [PubMed] [Google Scholar]
- Ishihama A., Mizumoto K., Kawakami K., Kato A., Honda A. Proofreading function associated with the RNA-dependent RNA polymerase from influenza virus. J Biol Chem. 1986 Aug 5;261(22):10417–10421. [PubMed] [Google Scholar]
- Jacobs L., Spaan W. J., Horzinek M. C., van der Zeijst B. A. Synthesis of subgenomic mRNA's of mouse hepatitis virus is initiated independently: evidence from UV transcription mapping. J Virol. 1981 Aug;39(2):401–406. doi: 10.1128/jvi.39.2.401-406.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong Y. S., Makino S. Mechanism of coronavirus transcription: duration of primary transcription initiation activity and effects of subgenomic RNA transcription on RNA replication. J Virol. 1992 Jun;66(6):3339–3346. doi: 10.1128/jvi.66.6.3339-3346.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo M., Makino S. Mutagenic analysis of the coronavirus intergenic consensus sequence. J Virol. 1992 Nov;66(11):6330–6337. doi: 10.1128/jvi.66.11.6330-6337.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassavetis G. A., Geiduschek E. P. RNA polymerase marching backward. Science. 1993 Feb 12;259(5097):944–945. doi: 10.1126/science.7679800. [DOI] [PubMed] [Google Scholar]
- Konings D. A., Bredenbeek P. J., Noten J. F., Hogeweg P., Spaan W. J. Differential premature termination of transcription as a proposed mechanism for the regulation of coronavirus gene expression. Nucleic Acids Res. 1988 Nov 25;16(22):10849–10860. doi: 10.1093/nar/16.22.10849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M. M., Baric R. S., Brayton P. R., Stohlman S. A. Characterization of leader RNA sequences on the virion and mRNAs of mouse hepatitis virus, a cytoplasmic RNA virus. Proc Natl Acad Sci U S A. 1984 Jun;81(12):3626–3630. doi: 10.1073/pnas.81.12.3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M. M., Brayton P. R., Armen R. C., Patton C. D., Pugh C., Stohlman S. A. Mouse hepatitis virus A59: mRNA structure and genetic localization of the sequence divergence from hepatotropic strain MHV-3. J Virol. 1981 Sep;39(3):823–834. doi: 10.1128/jvi.39.3.823-834.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M. M. Coronavirus: organization, replication and expression of genome. Annu Rev Microbiol. 1990;44:303–333. doi: 10.1146/annurev.mi.44.100190.001511. [DOI] [PubMed] [Google Scholar]
- Lai M. M., Patton C. D., Stohlman S. A. Replication of mouse hepatitis virus: negative-stranded RNA and replicative form RNA are of genome length. J Virol. 1982 Nov;44(2):487–492. doi: 10.1128/jvi.44.2.487-492.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. J., Shieh C. K., Gorbalenya A. E., Koonin E. V., La Monica N., Tuler J., Bagdzhadzhyan A., Lai M. M. The complete sequence (22 kilobases) of murine coronavirus gene 1 encoding the putative proteases and RNA polymerase. Virology. 1991 Feb;180(2):567–582. doi: 10.1016/0042-6822(91)90071-I. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibowitz J. L., Wilhelmsen K. C., Bond C. W. The virus-specific intracellular RNA species of two murine coronaviruses: MHV-a59 and MHV-JHM. Virology. 1981 Oct 15;114(1):39–51. doi: 10.1016/0042-6822(81)90250-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S., Joo M. Effect of intergenic consensus sequence flanking sequences on coronavirus transcription. J Virol. 1993 Jun;67(6):3304–3311. doi: 10.1128/jvi.67.6.3304-3311.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S., Joo M., Makino J. K. A system for study of coronavirus mRNA synthesis: a regulated, expressed subgenomic defective interfering RNA results from intergenic site insertion. J Virol. 1991 Nov;65(11):6031–6041. doi: 10.1128/jvi.65.11.6031-6041.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S., Keck J. G., Stohlman S. A., Lai M. M. High-frequency RNA recombination of murine coronaviruses. J Virol. 1986 Mar;57(3):729–737. doi: 10.1128/jvi.57.3.729-737.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S., Soe L. H., Shieh C. K., Lai M. M. Discontinuous transcription generates heterogeneity at the leader fusion sites of coronavirus mRNAs. J Virol. 1988 Oct;62(10):3870–3873. doi: 10.1128/jvi.62.10.3870-3873.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S., Stohlman S. A., Lai M. M. Leader sequences of murine coronavirus mRNAs can be freely reassorted: evidence for the role of free leader RNA in transcription. Proc Natl Acad Sci U S A. 1986 Jun;83(12):4204–4208. doi: 10.1073/pnas.83.12.4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S., Yokomori K., Lai M. M. Analysis of efficiently packaged defective interfering RNAs of murine coronavirus: localization of a possible RNA-packaging signal. J Virol. 1990 Dec;64(12):6045–6053. doi: 10.1128/jvi.64.12.6045-6053.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinkoth J., Wahl G. Hybridization of nucleic acids immobilized on solid supports. Anal Biochem. 1984 May 1;138(2):267–284. doi: 10.1016/0003-2697(84)90808-x. [DOI] [PubMed] [Google Scholar]
- Meulenberg J. J., de Meijer E. J., Moormann R. J. Subgenomic RNAs of Lelystad virus contain a conserved leader-body junction sequence. J Gen Virol. 1993 Aug;74(Pt 8):1697–1701. doi: 10.1099/0022-1317-74-8-1697. [DOI] [PubMed] [Google Scholar]
- Pachuk C. J., Bredenbeek P. J., Zoltick P. W., Spaan W. J., Weiss S. R. Molecular cloning of the gene encoding the putative polymerase of mouse hepatitis coronavirus, strain A59. Virology. 1989 Jul;171(1):141–148. doi: 10.1016/0042-6822(89)90520-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SantaLucia J., Jr, Kierzek R., Turner D. H. Stabilities of consecutive A.C, C.C, G.G, U.C, and U.U mismatches in RNA internal loops: Evidence for stable hydrogen-bonded U.U and C.C.+ pairs. Biochemistry. 1991 Aug 20;30(33):8242–8251. doi: 10.1021/bi00247a021. [DOI] [PubMed] [Google Scholar]
- Sawicki S. G., Sawicki D. L. Coronavirus minus-strand RNA synthesis and effect of cycloheximide on coronavirus RNA synthesis. J Virol. 1986 Jan;57(1):328–334. doi: 10.1128/jvi.57.1.328-334.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicki S. G., Sawicki D. L. Coronavirus transcription: subgenomic mouse hepatitis virus replicative intermediates function in RNA synthesis. J Virol. 1990 Mar;64(3):1050–1056. doi: 10.1128/jvi.64.3.1050-1056.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethna P. B., Hofmann M. A., Brian D. A. Minus-strand copies of replicating coronavirus mRNAs contain antileaders. J Virol. 1991 Jan;65(1):320–325. doi: 10.1128/jvi.65.1.320-325.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethna P. B., Hung S. L., Brian D. A. Coronavirus subgenomic minus-strand RNAs and the potential for mRNA replicons. Proc Natl Acad Sci U S A. 1989 Jul;86(14):5626–5630. doi: 10.1073/pnas.86.14.5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shieh C. K., Lee H. J., Yokomori K., La Monica N., Makino S., Lai M. M. Identification of a new transcriptional initiation site and the corresponding functional gene 2b in the murine coronavirus RNA genome. J Virol. 1989 Sep;63(9):3729–3736. doi: 10.1128/jvi.63.9.3729-3736.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shieh C. K., Soe L. H., Makino S., Chang M. F., Stohlman S. A., Lai M. M. The 5'-end sequence of the murine coronavirus genome: implications for multiple fusion sites in leader-primed transcription. Virology. 1987 Feb;156(2):321–330. doi: 10.1016/0042-6822(87)90412-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirai T., Go M. RNase-like domain in DNA-directed RNA polymerase II. Proc Natl Acad Sci U S A. 1991 Oct 15;88(20):9056–9060. doi: 10.1073/pnas.88.20.9056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaan W. J., Rottier P. J., Horzinek M. C., van der Zeijst B. A. Isolation and identification of virus-specific mRNAs in cells infected with mouse hepatitis virus (MHV-A59). Virology. 1981 Jan 30;108(2):424–434. doi: 10.1016/0042-6822(81)90449-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaan W. J., Rottier P. J., Horzinek M. C., van der Zeijst B. A. Sequence relationships between the genome and the intracellular RNA species 1, 3, 6, and 7 of mouse hepatitis virus strain A59. J Virol. 1982 May;42(2):432–439. doi: 10.1128/jvi.42.2.432-439.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaan W., Cavanagh D., Horzinek M. C. Coronaviruses: structure and genome expression. J Gen Virol. 1988 Dec;69(Pt 12):2939–2952. doi: 10.1099/0022-1317-69-12-2939. [DOI] [PubMed] [Google Scholar]
- Spaan W., Delius H., Skinner M., Armstrong J., Rottier P., Smeekens S., van der Zeijst B. A., Siddell S. G. Coronavirus mRNA synthesis involves fusion of non-contiguous sequences. EMBO J. 1983;2(10):1839–1844. doi: 10.1002/j.1460-2075.1983.tb01667.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto N., Kierzek R., Freier S. M., Turner D. H. Energetics of internal GU mismatches in ribooligonucleotide helixes. Biochemistry. 1986 Sep 23;25(19):5755–5759. doi: 10.1021/bi00367a061. [DOI] [PubMed] [Google Scholar]
- Yokomori K., Banner L. R., Lai M. M. Coronavirus mRNA transcription: UV light transcriptional mapping studies suggest an early requirement for a genomic-length template. J Virol. 1992 Aug;66(8):4671–4678. doi: 10.1128/jvi.66.8.4671-4678.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot R. J., ter Haar R. J., Horzinek M. C., van der Zeijst B. A. Intracellular RNAs of the feline infectious peritonitis coronavirus strain 79-1146. J Gen Virol. 1987 Apr;68(Pt 4):995–1002. doi: 10.1099/0022-1317-68-4-995. [DOI] [PubMed] [Google Scholar]
- de Groot R. J., van der Most R. G., Spaan W. J. The fitness of defective interfering murine coronavirus DI-a and its derivatives is decreased by nonsense and frameshift mutations. J Virol. 1992 Oct;66(10):5898–5905. doi: 10.1128/jvi.66.10.5898-5905.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Most R. G., Bredenbeek P. J., Spaan W. J. A domain at the 3' end of the polymerase gene is essential for encapsidation of coronavirus defective interfering RNAs. J Virol. 1991 Jun;65(6):3219–3226. doi: 10.1128/jvi.65.6.3219-3226.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Most R. G., Heijnen L., Spaan W. J., de Groot R. J. Homologous RNA recombination allows efficient introduction of site-specific mutations into the genome of coronavirus MHV-A59 via synthetic co-replicating RNAs. Nucleic Acids Res. 1992 Jul 11;20(13):3375–3381. doi: 10.1093/nar/20.13.3375. [DOI] [PMC free article] [PubMed] [Google Scholar]







