Abstract
Tus protein binds tightly to specific DNA sequences (Ter) on the Escherichia coli chromosome halting replication. We report here conditions for detecting the 1 : 1 Tus–Ter complex by electrospray ionization mass spectrometry (ESI-MS). ESI mass spectra of a mixture of Tus and nonspecific DNA showed ions predominantly from uncomplexed Tus protein, indicating that the Tus–Ter complex observed in the gas phase was the result of a specific interaction rather than nonspecific associations in the ionization source. The Tus–Ter complex was very stable using a spray solvent of 10 mM ammonium acetate at pH 8.0, and initial attempts to distinguish binding affinities of Tus and mutant Tus proteins for Ter DNA were unsuccessful. Increasing the ammonium acetate concentration in the electrospray solvent (800 mM at pH 8.0) increased the dissociation constants sufficiently such that relative orders of binding affinity for Tus and various mutant Tus proteins for various DNA sequences could be determined. These were in agreement with the dissociation constants determined in solution studies. A dissociation constant of 700 × 10−9 M for the binding of the mutant Tus protein A173T (where residue 173 is changed from alanine to threonine) to Ter DNA was estimated, compared with a value of ≤2 × 10−9 M for Tus where A173 was unchanged. This is the first example in which ESI-MS has been used to compare binding affinities of a DNA-binding protein with mutant proteins for specific DNA recognition sequences. It was also possible to estimate the strength of the interaction between Tus and a DNA sequence (TerH) that had been identified by database searching.
Keywords: Tus, DNA replication, electrospray ionization mass spectrometry, noncovalent complex, dis-sociation constant
Electrospray ionization mass spectrometry (ESI-MS) is now established as a powerful tool for analysis of the primary structure of biomolecules (Griffiths et al. 2001). More recently, this technique has been applied to the study of noncovalent biochemical complexes (Loo 1997). There are several technical difficulties to overcome to observe complexes involving macromolecules in the gas phase. For example, buffers used in solution studies of biochemical complexes are not usually volatile and therefore are not compatible with mass spectrometry. Furthermore, solvents used in ESI-MS typically contain an organic phase that could disrupt noncovalent complexes. Keeping this in mind, it is important to prepare the complex under conditions in which it maintains its native, folded state, and to use instrumental conditions such that it is ionized and transported to the mass analyzer intact. One of the most important criteria to be satisfied concerns whether noncovalent complexes observed in the gas phase reflect solution behavior or are the result of nonspecific associations in the ion source. As a consequence, ESI-MS of noncovalent complexes to date has been conducted on systems that have been well characterized in solution. Accumulation of information concerning a range of various binding partners prepared and analyzed under a range of conditions will shed light on the question of whether mass spectrometry can be used to study noncovalent interactions.
Interactions of proteins with nucleic acids are important in replication, repair, transcription, and translation. There have been fewer than 10 ESI-MS studies of noncovalent complexes of DNA with proteins and only a few of these involved complexes of intact proteins with double-stranded (ds)DNA (Cheng et al. 1996a; Potier et al. 1998; Craig et al. 1999). We have used ESI-MS to study the well-characterized interactions of Escherichia coli Tus protein (35,652 Daltons) with its DNA recognition sequence, TerB. Six termination sequences (TerA-F) have been identified on the E. coli chromosome, and each contains a consensus sequence that is ∼20 bp long. Tus binds as a monomer to termination sequences, halting replication. Termination of replication shows polarity in that when Tus binds to the chromosome, it stops the replication fork moving in one direction but not the other. A key to this polarity is found in the asymmetry of the complex revealed in the X-ray crystal structure of Tus with a 16-bp Ter DNA (Kamada et al. 1996). Equilibrium dissociation constants (Kd) for the Tus–Ter B complex have been measured using gel mobility shift and filter binding assays (Gottlieb et al. 1992; Skokotas et al. 1994; Coskun-Ari and Hill 1997) and in surface plasmon resonance (SPR) experiments (Neylon et al. 2000). The binding is very tight: Kd values are 3.3 × 10−13 M in buffer at pH 7.5, containing 150 mM glutamate (Gottlieb et al. 1992), and 0.5 × 10−9 M in buffer at pH 7.6, containing 250 mM KCl (Neylon et al. 2000). In the latter study, a range of KCl concentrations was investigated, and extrapolation of data to KCl = 150 mM gave Kd ∼1 × 10−12 M.
The availability of the X-ray crystal structure (Kamada et al. 1996), combined with in vivo and in vitro binding studies of native Tus compared with mutant proteins has enabled analysis of the relative contributions of various polar and hydrophobic interactions to binding. In addition, variant Ter sequences have been studied (Coskun-Ari and Hill 1997). In the Tus–Ter complex, the DNA lies in a positively charged cleft between N- and C-terminal domains that are joined by interdomain β strands (Kamada et al. 1996). Fourteen Tus residues make sequence-specific contacts with Ter DNA, and there are numerous polar contacts between Tus and the phosphate backbone. Importantly, on the side of the complex that allows the replication fork to proceed, one DNA strand makes extensive contact with Tus, whereas the other strand is predominantly exposed to solvent. In contrast, on the side of the complex in which replication is halted, Tus makes extensive contacts with both DNA strands (Kamada et al. 1996; Neylon et al. 2000).
In the cleft, the side chain of Ala 173 is involved in a hydrophobic interaction with the methyl group of a thymine base. When this alanine residue is changed to the more bulky threonine (A173T), Tus binds ∼4000-fold less tightly to TerB and is unable to halt replication in vivo (Skokotas et al. 1994; Neylon et al. 2000). Arg 198 lies just outside the core DNA-binding region on the side of the complex in which replication is halted and makes sequence-specific contacts with DNA (Kamada et al. 1996). Investigations of the interactions of these and other Tus mutants with Ter and nonspecific DNA and the effect of salt concentration on the binding have led to the proposal of a binding mechanism, wherein an initial nonspecific binding event involving interactions of Arg 198 and other basic residues with DNA is followed by sequence recognition by residues including Lys 89. Strong sequence specific contacts (e.g., with Ala 173) then can be made concomitant with a conformational change of Tus (Neylon et al. 2000).
Several ESI-MS studies of noncovalent complexes of DNA with proteins have been able to distinguish between binding of the protein with specific or nonspecific DNA (Cheng et al. 1996b; Potier et al. 1998), or in the case of a DNA repair protein, with damaged or undamaged DNA (Xu et al. 1999). In work reported here, we have studied the Tus–Ter interaction and describe the first use of ESI-MS to compare the relative strengths of binding of native and mutant proteins with specific DNA sequences. It was necessary in binding studies of the Tus–Ter complex using SPR to increase salt concentration to allow measurement of association and dissociation rates (and Kd). Similarly, in this work we needed to weaken the binding to distinguish between Tus and Tus mutants in complexes with Ter DNA; we used 800 mM ammonium acetate at pH 8.0, as the solvent in ESI-MS. In previous ESI-MS studies of DNA–protein complexes, low concentrations (usually 10 mM) of ammonium acetate or bicarbonate have been used. The use of an ESI time-of-flight mass spectrometer with a Z-spray probe has made the use of high salt possible. Conditions under which proteins are fully folded are more likely to be found if a wide range of salt conditions (10–2200 mM) are tested. The use of higher salt concentrations also decreases the chance that nonspecific gas phase associations of molecules will be observed (Sannes-Lowery et al. 2000).
Results and Discussion
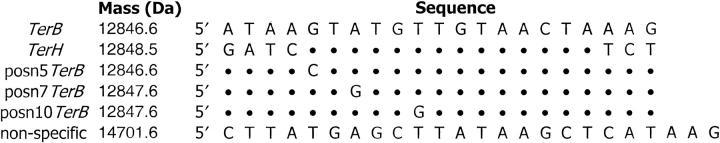
Figure 1 ▶ shows the sequences of DNA strands used in this work. All except the nonspecific DNA were 21 nucleotides long, and TerB had the same sequence used in our earlier SPR studies of Tus–Ter complexes (Neylon et al. 2000). In a previous study of the effect of base pair substitutions in Ter DNA on the Tus–Ter complex, Coskun-Ari and Hill (1997) used 33-bp oligonucleotides. The 21-bp oligonucleotides used in the present work contain the core sequence necessary for binding to Tus. The sequences termed position 5, 7, and 10 substitutions of TerB are the same variations termed position 6, 8, and 11 substitutions in the study by Coskun-Ari and Hill (1997). These sequences are here referred to as posn5TerB, posn7TerB, and posn10TerB, respectively. TerH was identified as a possible strong binding site for Tus by searching the E. coli genomic DNA sequence, but has not been examined previously by experiment (Coskun-Ari and Hill 1997). The nonspecific DNA is a self-complementary 24-bp sequence.
Fig. 1.
Sequences of dsDNAs used in this work; only one strand of each is shown. The dots indicate that the base in that position is the same as in the TerB sequence. The masses are those for double-stranded DNA, i.e., the strand shown in addition to its complement. Da, Dalton.
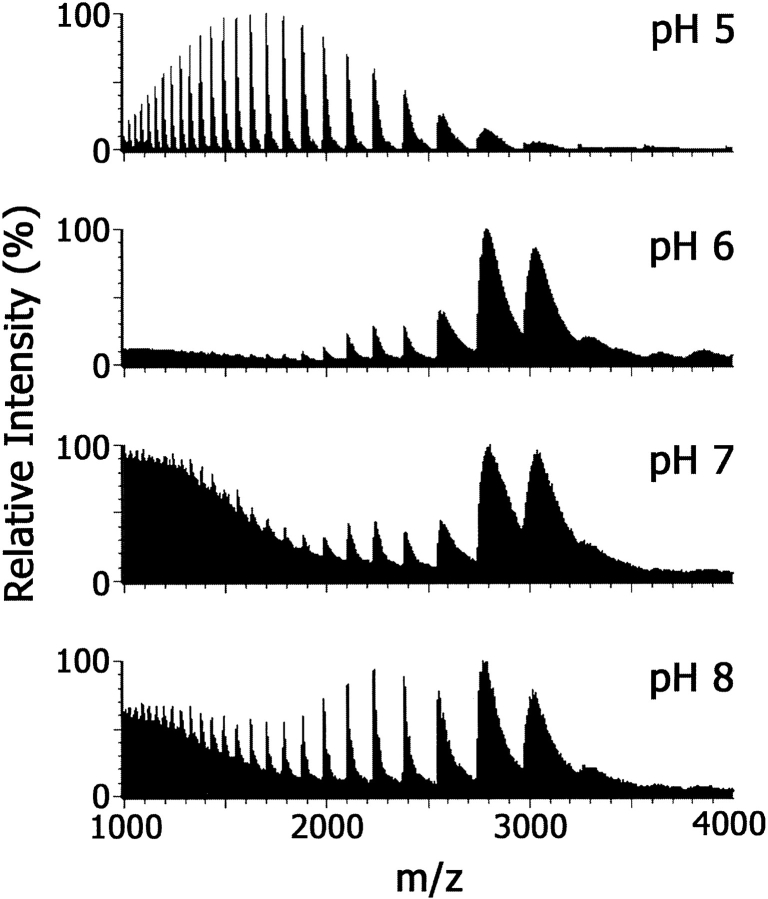
As a first step in ESI-MS of noncovalent DNA–protein complexes, it is important in both the preparation of the complex and in choosing the spray solvent to use solution conditions in which the protein is in its native, folded conformation. Figure 2 ▶ shows ESI mass spectra of Tus that had been dialyzed into 10 mM ammonium acetate (NH4OAc) over the pH range 5.0 to 8.0 and infused directly into the ionization source. The ESI mass spectrum of Tus at pH 5.0 showed numerous ions with [M + 25H]25+ (m/z 1427.1) the most abundant. At higher pH values, the charge distribution is markedly different with ions observed at higher values of m/z with [M + 13H]13+ (m/z 2743.5) the most abundant. This change in the charge envelope generally is observed on acidification of proteins and has been explained in terms of unfolding of the protein leading to exposure of a greater number of basic residues (Konermann and Douglas 1998; Jarrold 1999). There is evidence from nuclear magnetic resonance spectroscopy that Tus is fully folded at pH 8.0, but unfolds at pH <6.0 (G. Otting and N.E. Dixon, unpubl.). At pH ≥6.0, the ions are very broad (m/z ∼250 at half height), indicating incomplete desolvation of the protein under these conditions. This observation suggests that water/salt molecules trapped in the folded structure are released as the protein unfolds.
Fig. 2.
ESI mass spectra of Tus (10 μM) in 10 mM NH4OAc at pH values of 5.0, 6.0, 7.0, and 8.0. Instrumental conditions were as described in Materials and Methods except that the desolvation temperature was 60°C.
In these preliminary ESI-MS experiments, low desolvation temperatures (60°C) were used. In previous work, low temperatures had enabled detection of noncovalent complexes of intercalators with double-stranded DNA (Kapur et al. 1999). These conditions gave spectra of the Tus–Ter complex in which the peaks were broad (Fig. 2 ▶, pH 6.0 to 8.0), and mass accuracy therefore was low. An experiment was conducted to determine the effect of desolvation temperature on ESI mass spectra of a 1 : 1 Tus–TerB complex in 10 mM NH4OAc at pH 8.0, over the range 60 to 240°C (data not shown). As expected, increasing desolvation temperature resulted in sharper peaks in the ESI mass spectra. At 240°C, the complex remained intact and sharp peaks were obtained with the highest signal-to-noise ratio. The width-at-half-height of ions from the complex was m/z of ∼5 (m/z ∼40 at base). The signal-to-noise ratio was improved by the use of an elevated pressure of argon in the collision cell and a collision energy of ∼20 eV. This presumably reduces the energy spread of the ions entering the time-of-flight analyzer.
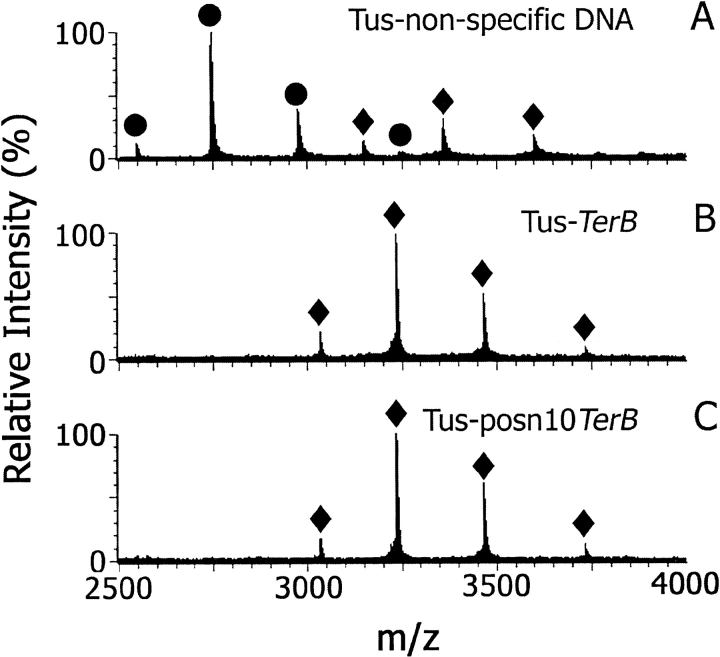
Figure 3 ▶ shows ESI mass spectra of mixtures of Tus with nonspecific (Fig. 3A ▶), TerB (Fig. 3B ▶), and posn10TerB (Fig. 3C ▶) DNA in 20 mM ammonium acetate at pH 8.0, under optimized instrumental conditions. For samples containing TerB and posn10TerB, the only significant ions in the spectra were from 1 : 1 Tus–DNA complexes ([M + 14H]14+, [M + 15H]15+, and [M + 16H]16+ ions, at m/z 3465.5, m/z 3234.5, and m/z 3032.4, respectively; see Table 1). Significantly, in the spectrum of Tus with nonspecific DNA recorded under the same conditions, the predominant ions were from free Tus (Fig. 3A ▶). The observation of the complex of Tus with TerB DNA but not with nonspecific DNA suggests that the complex observed in the gas phase is not the result of nonspecific associations in the ionization source. There is also a small amount of Tus-nonspecific DNA complex evident in the spectrum. Weak binding to nonspecific DNA also was observed in solution studies with Tus (Coskun-Ari and Hill 1997; Neylon et al. 2000) and in studies on other DNA-binding proteins (Ha et al. 1992). Furthermore, the first step in the proposed binding mechanism of Tus to TerB involves nonspecific electrostatic interactions of positively charged residues of Tus with the phosphate backbone of DNA (Neylon et al. 2000).
Fig. 3.
ESI mass spectra of Tus–dsDNA complexes (10 μM) in 20 mM NH4OAc at pH 8.0. (A) Tus-nonspecific DNA, (B) Tus–TerB, (C) Tus–posn10TerB. (circles) Ions from free Tus protein; (diamonds) ions from Tus–dsDNA complexes.
Table 1.
Calculated values of m/z for ions observed in ESI mass spectra
| Protein | Complexa | [M + 11H]11+ | [M + 12H]12+ | [M + 13H]13+ | [M + 14H]14+ | [M + 15H]15+ | [M + 16H]16+ |
| Native Tus | Freeb | 3242.1 | 2972.0 | 2743.5 | 2547.6 | 2377.8 | 2229.3 |
| (35 652 Da) | TerBc | 4410.0 | 4042.5 | 3731.7 | 3465.2 | 3234.2 | 3032.2 |
| Nonspecific | 4578.6 | 4197.1 | 3874.4 | 3597.7 | 3357.9 | 3148.1 | |
| His6Tus | Freeb | 3340.7 | 3062.4 | 2826.9 | 2625.1 | 2450.1 | 2297.1 |
| (36 737 Da) | TerBc | 4508.6 | 4133.0 | 3815.2 | 3542.7 | 3306.6 | 3100.0 |
| Nonspecific | 4677.3 | 4287.6 | 3957.8 | 3675.2 | 3430.3 | 3215.9 | |
| R198A | Freeb | 3333.0 | 3055.3 | 2820.4 | 2619.0 | 2444.5 | 2291.8 |
| (36 652 Da) | TerBc | 4500.9 | 4125.9 | 3808.6 | 3536.6 | 3300.9 | 3094.7 |
| Nonspecific | 4669.5 | 4280.5 | 3951.3 | 3669.1 | 3424.6 | 3210.6 | |
| A173T | Freeb | 3343.5 | 3064.9 | 2829.2 | 2627.2 | 2452.1 | 2298.9 |
| (36 767 Da) | TerBc | 4511.3 | 4135.5 | 3817.4 | 3544.8 | 3308.6 | 3101.8 |
| Nonspecific | 4680.0 | 4290.1 | 3960.1 | 3677.3 | 3432.3 | 3217.8 |
a This column shows the DNA present in the complex.
b "Free" refers to the protein in the absence of DNA.
c The m/z values for complexes of Tus proteins with posn5,7,10TerB are not given since they are the same within 0.2 Da as the complexes with TerB DNA.
The posn10TerB (a T • A base pair changed to G • C) was shown using filter binding assays to bind to Tus with an equilibrium dissociation constant (Kobs) of 1204 × 10−13 M compared with the value for native TerB of 9 × 10−13 M. In addition, the in vivo replication arrest activity of Tus bound at a native TerB site was 95%, compared with only 2% efficiency for this substituted DNA (Coskun-Ari and Hill 1997). These observations were explained in terms of removal of a hydrophobic interaction between Val 234 and the thymidine in the T • A base pair normally present at position 10 of TerB (Coskun-Ari and Hill 1997). Under the conditions of this ESI-MS experiment, we could not distinguish between binding of Tus with TerB (Fig. 3B ▶) or posn10TerB (Fig. 3C ▶). This is in accord with the relative concentrations of posn10TerB–Tus complex (Tuscomplex) and free Tus (Tusfree) in solution, calculated using the value of Kobs (1204 × 10−13 M, above): [Tusfree = 0.03 μM, Tuscomplex = 9.97 μM. Thus, if binding in the gas phase under our experimental conditions were at least as tight as in solution, then ions from free Tus would not be readily observable in the ESI mass spectrum. This also assumes that the response factors of Tus and Tus–TerB complex are comparable. The response factor is used to describe the efficiency with which gas phase ions are formed and detected in the mass spectrometer. Certainly, given the lower net charge on the complex than on free Tus, it seems unlikely that in positive ion ESI mass spectra the response factor would be lower for free Tus than for the DNA–Tus complex.
Similarly, in ESI mass spectra of complexes of unmodified Tus with posn5TerB, posn7TerB, or with TerH, there were no ions from free Tus or DNA. Furthermore, mixtures of TerB with Tus, the A173T mutant of Tus, or N-terminal (His)6-tagged Tus (his6Tus) all gave ESI mass spectra in which only ions from complexes and not from free binding partners were observed. This suggests that under the conditions of these experiments, all these complexes were too tightly bound to enable observation of free binding partners. In SPR studies, the equilibrium dissociation constants for Tus and his6Tus were indistinguishable (Neylon et al. 2000). In subsequent experiments (below), his6Tus (rather than unmodified Tus) was used to enable more direct comparisons with mutant Tus proteins (A173T and R198A), which both carried a hexahistidine tag.
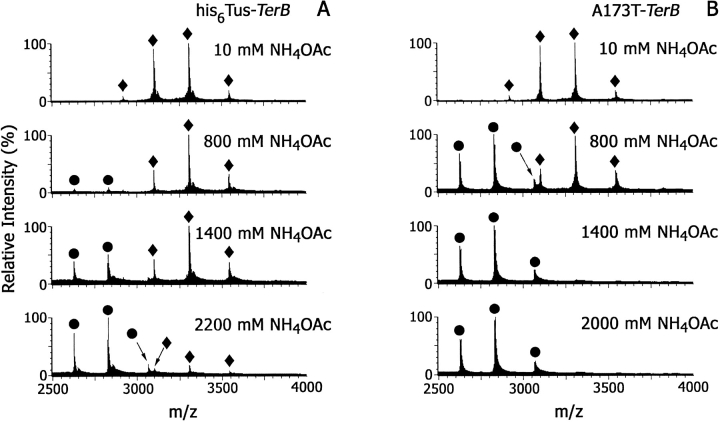
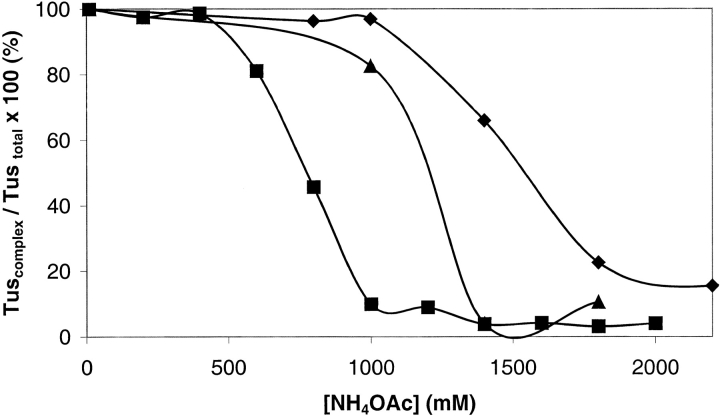
In SPR experiments with Tus or his6Tus, it was possible to determine equilibrium dissociation constants only when the binding interaction was weakened by increasing the KCl concentration (Neylon et al. 2000). Similarly, we reasoned that increasing the ionic strength of the spray solvent would allow distinction between complexes of Tus and mutants with DNA in ESI mass spectra. Most ESI-MS studies of noncovalent complexes have used 10–50 mM ammonium acetate, with one report of a mass spectrum of dsDNA obtained in 150 mM ammonium acetate at pH 7.0 (Hofstadler and Griffey 2001). An experiment was conducted in which ESI-MS spectra of Tus–Ter (10 μM) complexes were acquired over a range of ammonium acetate concentrations from 10 to 2200 mM at pH 8.0. Figure 4 ▶ shows the effect of increasing NH4OAc concentration on the his6Tus–TerB and A173T–TerB complexes. Ions were observed at m/z 3543.2, 3307.0, and 3100.4 for his6Tus–TerB, and at m/z 3545.5, 3309.2, and 3102.4 for A173T–TerB. The A173T–TerB complex is almost completely dissociated when the solvent is 800 mM NH4OAc, whereas the ESI mass spectrum of the his6Tus–TerB complex at this salt concentration shows ions only from the complex. The latter complex is ∼50% dissociated at NH4OAc concentration = 1400 mM at pH 8.0 and is not completely dissociated until NH4OAc concentration ≥2200 mM.
Fig. 4.
ESI mass spectra of the Tus–dsDNA complexes (10 μM). (A) his6Tus–TerB and (B) A173T–TerB at NH4OAc concentration = 10–2200 mM. (circles) Ions from free protein; (diamonds) ions from protein–dsDNA complex.
In all experiments, as ions corresponding to free Tus increased, ions appeared that corresponded to both single strands and dsTer DNA (see electronic supplemental material). These additional ions were at m/z 2119.7 and 1590.4 ([M + 3H]3+ and [M + 4H]4+ of one Ter strand), at m/z 1624.7 ([M + 4H]4+ of the other Ter strand), and at m/z 2142.6 and 1836.9 ([M + 6H]6+ and [M + 7H]7+ of dsTer). The appearance of the DNA as single strands when it dissociates from Tus is expected based on other work in this laboratory showing that dsDNA denatures as the desolvation temperature is increased above 60°C. It seems likely that the response factor for DNA would be different from that for free Tus protein or the Tus–Ter complexes. Therefore, we have compared intensities of ions from free Tus with intensities of ions from complex in the following experiments.
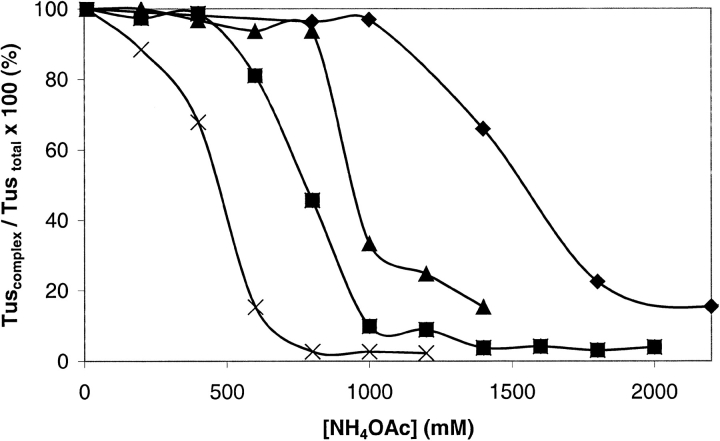
An experiment was conducted over the same range of salt concentrations used to obtain the data in Figure 4 ▶, but comparing Tus–TerB with his6Tus–TerB, A173T–TerB, and R198A–TerB. No significant differences could be detected between ESI mass spectra of Tus–TerB and his6Tus–TerB at any salt concentration in the range 10 to 2200 mM (data not shown). This is in agreement with the SPR studies (Neylon et al. 2000). Figure 5 ▶ compares the complexes his6Tus–TerB, A173T–TerB, and R198A–TerB. The data were obtained by summing the intensities of all ions from the complex (Tuscomplex) and from free Tus (Tusfree) and expressing each as a percentage of Tustotal (Tustotal = Tusfree + Tuscomplex; data shown for Tuscomplex/Tustotal). The amount of each of the complexes decreased with increasing NH4OAc concentration. The relative order of binding affinities can be determined by comparing the NH4OAc concentration at which each complex is 50% dissociated: his6Tus > R198A > A173T. The solution Kd values measured in 250 mM KCl using SPR for the his6Tus–TerB, R198A–TerB, and A173T–TerB complexes were 0.5 × 10−9, 130 × 10−9, and 2000 × 10−9 M, respectively (Neylon et al. 2000). The present data are in very reasonable agreement.
Fig. 5.
Stability of complexes of his6Tus and Tus mutants with TerB. The data show the decreasing amounts of Tus (or mutants) in the complex with dsDNA (Tuscomplex) as a percentage of the total amount of Tus (Tustotal), as a function of NH4OAc concentration. These values were determined by summing the intensities of all ions from Tusfree and all ions from Tuscomplex. Tusfree + Tuscomplex = Tustotal. (diamonds) his6Tus–TerB, (triangles) R198A–TerB, (squares) A173T–TerB.
Figure 6 ▶ shows data for the complexes of TerB and posn10TerB with each of his6Tus and A173T. The order of stability of these complexes in increasing ammonium acetate concentrations is his6Tus–TerB > his6Tus–posn10TerB > A173T– TerB > A173T–posn10TerB. The dissociation constants of Tus–TerB and Tus–posn10TerB complexes are reported to be 9 × 10−13 and 1204 × 10−13 M, respectively (Coskun-Ari and Hill 1997). The A173T–TerB complex has not been compared previously with the A173T–posn10TerB complex. It might be expected that changing both an important sequence specific contact in the protein (A173T) concomitant with changing a base in the DNA causes the binding to be less avid than changing either one of the binding partners on its own.
Fig. 6.
Stability of complexes of his6Tus and A173T with TerB and posn10TerB. The data show the decreasing amounts of his6Tus or A173T in the complex with dsDNA (Tuscomplex) as a percentage of the total amount of Tus (Tustotal), as a function of NH4OAc concentration. (diamonds) his6Tus–TerB, (triangles) his6Tus–posn10TerB, (squares) A173T–TerB, (multiplication symbols) A173T–posn10TerB.
These relative binding affinities were confirmed in competition experiments conducted in 800 mM ammonium acetate at pH 8.0. ESI mass spectra of 1 : 1 : 1 mixtures (10 μM each) of various protein and DNA samples were acquired. For example, to determine whether his6Tus, A173T, or R198A bind more tightly to TerB, the following mixtures were set up: his6Tus : A173T : TerB, his6Tus : R198A : TerB, and R198A : A173T : TerB. Table 2 summarizes the competition mixtures that were used and shows the complexes and free binding partners that were observed in the spectra. Under the conditions of these experiments, the Kd values of the complexes compared here were sufficiently disparate that each ESI mass spectrum showed ions from only one DNA–protein complex together with ions from the free protein not involved in the complex. Analysis of the spectra summarized in Table 2, part A, shows clearly that the proteins bind to TerB in the order his6Tus > R198A > A173T. This is consistent with data in Figure 5 ▶ and Kd values measured in SPR experiments. Analysis of the spectra summarized in Table 2, part B, show that the proteins bind to posn10TerB in the same order.
Table 2.
Results of competition experiments
| Ions observed in the ESI-mass spectrum | ||||||
| Mixture components | His6Tus | A173T | R198A | His6Tus-TerB | A173T-TerB | R198A-TerB |
| A | ||||||
| His6Tus: A173T: TerB | × | √ | — | √ | × | — |
| His6Tus: R198A: TerB | × | — | √ | √ | — | × |
| R198A: A173T: TerB | — | √ | × | — | × | √ |
| B | His6Tus | A173T | R198A | His6Tus- posn10TerB | A173T- posn10TerB | R198A- posn10TerB |
| His6Tus: A173T: posn10TerB | × | √ | — | √ | × | — |
| His6Tus: R198A: posn10TerB | × | — | √ | √ | — | × |
| R198A: A173T: posn10TerB | — | √ | × | — | × | √ |
Mixture components were present in 1:1:1 molar ratios in 800 mM NH4OAc, pH 8. In the ESI mass spectra of these mixtures, only one complex was observed: (×) ions not observed; (√) ions observed; (—) not a mixture component in this experiment.
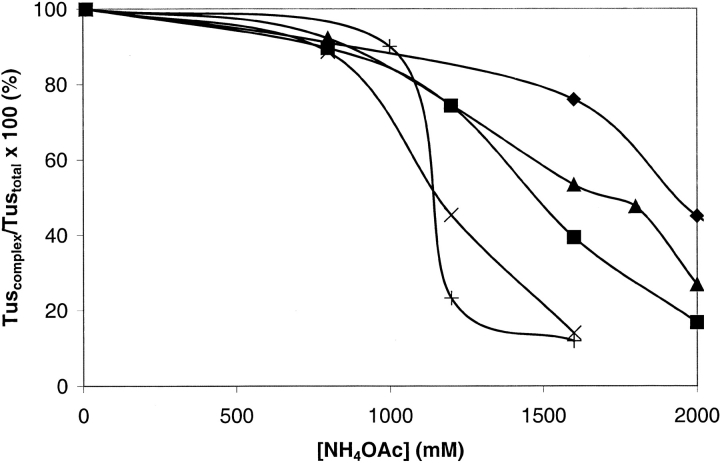
The relative binding affinities of his6Tus for TerB DNA substituted at positions 5, 7, and 10 and for TerH DNA could not be determined in competition experiments because the difference in mass between an A • T and a G • C base pair is only 1 Dalton, and the resolution in the ESI mass spectra was not sufficient to distinguish between complexes involving these different DNA sequences. The relative stabilities of these complexes were tested in NH4OAc in the same way as experiments shown in Figures 5 and 6 ▶ ▶. The data in Figure 7 ▶ show that the modified Ter sequences bind to his6Tus in the order TerB > posn5TerB > TerH > posn7TerB ∼ posn10TerB. These results are consistent with values of Kobs measured in solution of 9 × 10−13, 16 × 10−13, 139 × 10−13, and 1204 × 10−13 M for TerB, posn5TerB, posn7TerB, and posn10TerB, respectively (Coskun-Ari and Hill 1997). It is difficult to determine unequivocally from these data the position in the binding order of posn10TerB relative to posn7TerB. In posn5TerB, a G • C has been changed to a C • G base pair. This removes an interaction between Arg 198 of Tus and the N-3 atom of the guanine residue. In posn7TerB, an A • T base pair has been changed to G • C, removing interactions of the O-2 of the thymine base with Lys 89, and a major groove interaction of the thymine methyl group with Thr 139 (Kamada et al. 1996; Coskun-Ari and Hill 1997). The TerH site was identified by database searching (Coskun-Ari and Hill 1997). On the basis of Ter base pair substitution studies, it was proposed to be a moderately strong site, but this was not confirmed by experiment. The present results suggest that this is true.
Fig. 7.
Stability of complexes of his6Tus with the variant Ter DNAs. The data show the decreasing amounts of his6Tus in the complex with dsDNA (Tuscomplex) as a percentage of the total amount of Tus (Tustotal), as a function of NH4OAc concentration. (diamonds) his6Tus–TerB, (triangles) his6Tus–posn5Ter, (squares) his6Tus–TerH, (multiplication symbols) his6Tus–posn7TerB, (plus signs) his6Tus–posn10TerB.
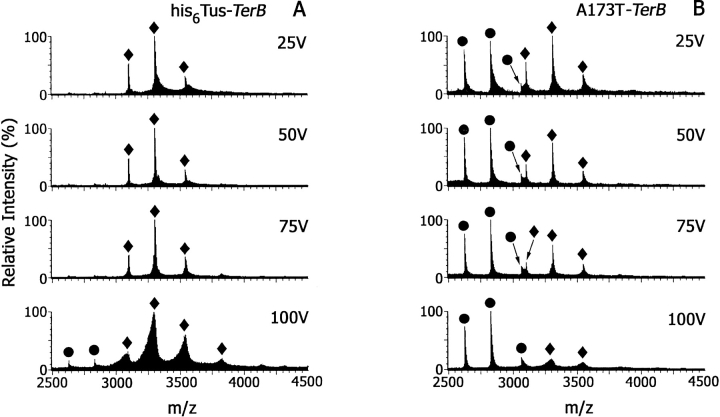
Collision-induced dissociation (CID) experiments (either in the source or collision cell) have been used in ESI-MS studies of noncovalent complexes as a measure of the stability of the binding interaction (Schwartz et al. 1995; Wan et al. 2000). In a study of the interactions of the Trp repressor with its consensus operator DNA sequence, an increase in cone voltage from 100 to 200 V resulted in only partial dissociation of the complex with the appearance of low abundance ions at values of m/z corresponding to complexes with a mass ∼150 Daltons lower than ions from intact complex. These ions were thought to arise from depurination of DNA (Potier et al. 1998). In our early experiments, attempts to disrupt the Tus–TerB or A173T–TerB complexes prepared and analyzed in 10 mM ammonium acetate at pH 8.0, by increasing the cone voltage from 50 to 100 V were unsuccessful. Furthermore, none of the complexes of Tus with substituted Ter sequences (Fig. 1 ▶) or of A173T with any of these DNA sequences was dissociated under these conditions. This experiment was repeated using 800 mM ammonium acetate at pH 8.0 as solvent. Figure 8 ▶ shows ESI mass spectra of the his6Tus–TerB (Fig. 8A ▶) and A173T–TerB (Fig. 8B ▶) complexes, as a function of cone voltage. At 25 V, A173T–TerB is ∼50% dissociated in this solvent, and the complex further dissociates as the cone voltage is increased to 100 V. In contrast, the his6Tus–TerB complex remains intact up to 100 V. This CID experiment confirms the data above which show that his6Tus binds the TerB sequence more tightly than the mutant A173T. The spectrum of the his6Tus–TerB complex (Fig. 8A ▶) obtained using a cone voltage of 100 V shows that ions from the complex are shifted to lower m/z values. Tus protein alone was stable at this cone voltage, suggesting that as in experiments with the Trp repressor (Potier et al. 1998) there may have been some fragmentation of DNA in the complex. In a separate experiment, resolution was sufficient to enable observation of an ion corresponding to a mass loss of ∼134 Daltons (data not shown), consistent with loss of adenine.
Fig. 8.
The effect of increasing cone voltage on ESI mass spectra of (A) his6Tus–TerB and (B) A173T–TerB, each 10 μM in 800 mM NH4OAc at pH 8.0. (circles) Ions from free protein; (diamonds) ions from protein–dsDNA complex.
The Tus–Ter interaction involves many electrostatic contacts, therefore increasing salt concentration markedly decreases the stability of the Tus–TerB complex in solution studies. This general observation has been confirmed in the present ESI mass spectra. The solvent used in the SPR study was 50 mM Tris-HCl at pH 7.5, 0.1 mM EDTA, 0.1 mM DTT, 0.005% Nonidet P-20, 250 mM KCl, which is markedly different from ammonium acetate used for ESI-MS. To comment further on whether the solution and gas phase complexes are the same, values of Kd need to be measured by titrating Tus with TerB and comparing these values with those determined in solution at similar ionic strength. The delineation of solvent and ESI-MS instrumental conditions that allow discrimination among the relative stabilities of various complexes is a starting point for being able to measure Kd values by ESI-MS.
ESI-MS has some advantages over filter binding and gel retardation assays for study of protein–DNA interactions in that these techniques necessarily require separation of bound and free mixture components for analysis and this may perturb the equilibrium position (Hagmar et al.1995). However, there are also complicating issues in determination of Kd values by ESI-MS. The first, alluded to above, is that the relative intensities of gas phase ions from free binding partners and complex may not correspond to the relative amounts of these species in solution if the response factors are markedly different. Response factors include the relative ionization efficiencies of solution components, but their determination is not a simple matter of predicting ions based on solution pKa values (Wang and Cole 1994; Constantopoulos et al. 1999; Cech and Enke 2000). The relative response factors for Tus and Tus–Ter complexes were determined by titrating A173T into a solution containing 1 : 1 his6Tus–TerB complex in 800 mM NH4OAc at pH 8.0. Under these conditions, the his6Tus–TerB complex is stable and A173T is not expected to bind TerB (Fig. 4 ▶). The total concentration of all Tus (Tustotal; A173T + his6Tus in the complex) was maintained at 10 μM. A plot of the ratio of the intensities of ions from A173T (Tusfree) to the intensities of ions from the complex (Tuscomplex) in the ESI mass spectrum against the ratios of free A173T and his6Tus–TerB complex added to the solution showed the response factors of free Tus and the complex to be the same within experimental error (see electronic supplemental material). Note that relative orders of binding obtained by determining the NH4OAc concentration at which complexes dissociate (Figs. 4–7 ▶ ▶ ▶ ▶) are independent of response factors of free Tus and complex.
A second complication is that electrostatic interactions are thought to be strengthened in vacuo. Therefore, the relative contributions of electrostatic interactions, hydrogen bonding, and hydrophobic and van der Waals interactions to the free energy of binding will influence stabilities of noncovalent complexes in the ESI source (Loo 1997). An example of where this may have an impact on values of dissociation constants estimated in the gas phase would be in comparisons of Tus with R198A. Arg 198 is involved in interactions with the negatively charged DNA backbone. If this and other electrostatic interactions are strengthened in the mass spectrometer, then the difference in the strength of TerB binding by Tus and R198A would be greater in the gas phase than in solution.
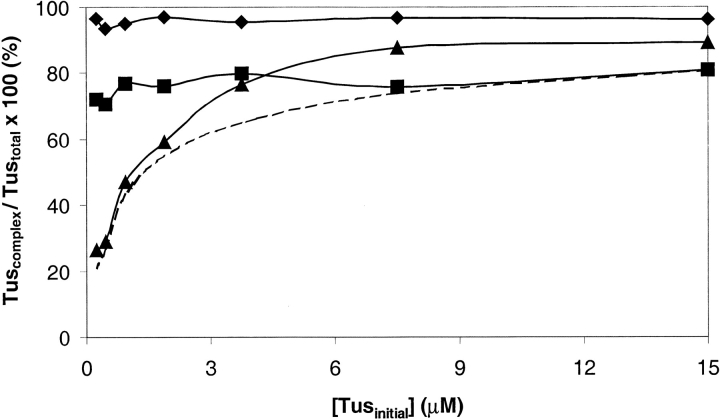
Scatchard plots (for measuring Kd) determined in the gas phase have been generated for noncovalent complexes of vancomycin antibiotics with tripeptides (Lim et al. 1995), and for complexes of aminoglycoside antibiotics with RNA (Sannes-Lowery et al. 2000), and were in reasonable agreement with solution data. We attempted to titrate Tus proteins with TerB DNA but encountered a small amount of precipitation of a component of the solution at stoichiometries of TerB : Tus <1 : 1. Therefore, to estimate values of the Kd of Tus–TerB complexes, we collected ESI-MS spectra on serial twofold dilutions of the 1 : 1 complexes (over the range 0.23–15 μM in 800 mM ammonium acetate at pH 8.0). The results are presented in Figure 9 ▶. For the his6Tus and R198A complexes, there was no change in the relative amounts of complex and free Tus as the complexes were diluted (Fig. 9 ▶). However, for the A173T–TerB complex, dilution gave rise to changes in concentrations of these species that suggest that under the conditions of this experiment, Kd for the A173T–TerB complex is ∼700 × 10−9 M (Fig. 9 ▶). For his6Tus–TerB and R198A–TerB, inability to observe an effect of dilution on relative amounts of free protein and complex implies that values of Kd of these complexes are ≤2 × 10−9 M, based on the assumption that ions from free Tus can be observed once their total intensity (Tusfree) is ∼6% of that of all Tus ions (Tustotal) in the ESI mass spectrum. For comparison, the solution Kd values measured in 250 mM KCl using SPR for the A173T– TerB, his6Tus–TerB, and R198A–TerB complexes are 2000 × 10−9, 0.5 × 10−9, and 130 × 10−9 M, respectively (Neylon et al. 2000).
Fig. 9.
The data show the amounts of Tus (or mutants) in the complex with TerB in 800 mM NH4OAc at pH 8.0 (Tuscomplex), as a percentage of the total amount of Tus (Tustotal) plotted against [Tus]initial. The intensities of all ions in the complex were summed giving Tuscomplex. The intensities of all ions from free Tus (Tusfree) and from the complex (Tuscomplex) were summed to give Tustotal. (diamonds) his6Tus–TerB, (squares) R198A–TerB, (triangles) A173T–TerB. (dashed line) The calculated curve for dissociation constant = 700 × 10−9 M.
One of the primary experimental considerations in gas phase detection of noncovalent complexes of biological molecules is choice of conditions that are sufficiently gentle to maintain the integrity of the complex. In our study of the Tus–TerB interaction, we were able to observe ESI mass spectra of the complex with high sensitivity: down to 0.23 μM (11.5 pmole in 50 μL). The specificity of this interaction was shown by the inability to observe significant amounts of a complex with nonspecific DNA under conditions in which the Tus–Ter complexes were very stable. To distinguish subtle changes in dissociation constants using ESI-MS, it was necessary to weaken the binding by increasing the NH4OAc concentration in the spray solvent. The relative orders of binding affinity determined were in reasonable agreement with solution studies, suggesting that the complexes observed in ESI mass spectra are representative of those in solution. This confirms that a specific interaction was being observed. Once conditions under which titrations of Tus with TerB can be conducted without precipitation of mixture components, it will be possible to determine dissociation constants and to make more direct comparisons with values measured in solution.
The question as to the validity of dissociation constants measured in the gas phase has been addressed (Lim et al. 1995; Sannes-Lowery et al. 2000). The ionization process itself may perturb the structure of the complex and therefore the equilibrium position. The electrospray ionization process requires desolvation of charged droplets. In DNA–protein interactions, water molecules may have an integral structural role, for example, in forming H-bonds within and between binding partners (Schwabe 1997). The loss of water from the complex may have important implications for comparisons between the gas phase and solution. In the Tus–TerB complex, water molecules are involved in H-bonding between the DNA and protein (Kamada et al. 1996). Once DNA–protein contacts are formed in solution, whether any effects of instrumental conditions will be observed depends on the difference between the time required for a conformational change of the complex and the time between desolvation and detection by the mass analyzer. The extent of effects caused by transferal to the gas phase will depend on the nature of the interactions holding the complex together. The possible strengthening of electrostatic interactions in vacuo will have a greater impact on complexes in which these interactions have a dominant role in stability of the complex. Studies by ESI-MS of a range of extensively characterized complexes are important to establish guidelines for the magnitude of such effects. It might be possible to measure the relative contribution of a particular interaction to stability of a complex by comparing the relative effects of various mutations on gas and solution phase dissociation constants. For example, mutation of a residue involved in an electrostatic interaction might have a greater effect on dissociation constants in the gas phase than in solution.
Materials and methods
All reagents were of analytical grade. MilliQ water was used in all experiments. Oligonucleotides (Fig. 1 ▶) were obtained from GeneWorks (Adelaide, South Australia) as the trityl on derivatives. They were deprotected using standard procedures and purified by two stages of reverse-phase high pressure liquid chromatography as described previously (Wickham et al. 1995). Concentrations of single-stranded oligonucleotides were estimated by measurement of ultraviolet light absorbance at 260 nm using values of ɛ260 for adenine, guanine, cytosine, and thymine of 15,400, 11,700, 7300, and 8300 M−1 cm−1, respectively (Sambrook et al. 1989).
Characterization of proteins
Unmodified Tus, his6Tus, A173T (his6Tus in which Ala 173 was changed to Thr), and R198A (his6Tus in which Arg 198 was changed to Ala) were expressed in E. coli, purified, and stored as previously described (Neylon et al. 2000). These protein samples had been characterized previously by mass spectrometry (Neylon et al. 2000), giving masses in agreement with calculated values (Table 1). Tus concentrations were determined by measurement of ultraviolet light absorbance at 280 nm, using ɛ280 = 39,700 M−1 cm−1 (Coskun-Ari et al. 1994).
Preparation of double-stranded (ds)DNA
Complementary single-stranded oligonucleotides (2.5 mM in 0.1 M ammonium acetate at pH 8.0) were heated to ≥20°C above melting temperature and allowed to cool slowly overnight. Annealed DNA was stored at 4°C before use.
Preparation of DNA–protein complexes
In first attempts to prepare a Tus–Ter complex, Tus and annealed TerB DNA were mixed in a 1 : 1 molar ratio and dialyzed together against 10 mM ammonium acetate at pH 8.0. This resulted in ESI mass spectra that showed a mixture of ions from free Tus and the complex. In addition, a small amount of precipitate was observed in dialyzed samples. Subsequently, the complex was prepared by first dialyzing Tus (1–15 μM) against 10 mM ammonium acetate at pH 8.0, at 4°C, followed by mixing it with an equimolar amount of dsDNA (typically 500 μL of protein to 1 μL of DNA in 0.1 M NH4OAc). The mixture was left on ice for 1.5 h before injection into the mass spectrometer. In experiments in which NH4OAc concentration was varied, a small volume of 10 M ammonium acetate at pH 8.0 was added to the mixture 1 h before mass spectrometry. In competition experiments, the two Tus protein samples were mixed in 800 mM NH4OAc at pH 8.0 and allowed to equilibrate for 30 min, followed by addition of an equimolar amount of dsDNA, giving a final concentration of each component of the mixture of 10 μM. The mixtures were left on ice for 1.5 h before direct injection into the mass spectrometer. ESI mass spectra of samples of free proteins used in these mixtures were acquired just before and after ESI mass spectra of mixtures to ensure that there had been no drift in calibration.
Electrospray ionization mass spectrometry
ESI mass spectra were acquired using a Qtof2 mass spectrometer (Micromass, Wyntheshawe, UK) equipped with a Z-spray electrospray ionization source. This spectrometer has an m/z range of 10,000. Samples were injected directly into the source using a Harvard Model 22 syringe pump at flow rates between 5 and 10 μL min−1. The best conditions for obtaining mass spectra of the DNA–protein complex were capillary, 2.5 kV; cone, 50 V; source block temperature, 40°C; desolvation temperature, 240°C; collision energy, 20 eV; aperture, 13; and transport, 6. Spectra were acquired in positive ion mode over a m/z range of 1000–7000. Typically, 25–30 scans were summed to give representative spectra. The data were calibrated against a standard CsI solution (750 μM) over the same m/z range. The ESI spectra shown in this work and those used for measurements of intensities of ions were not subjected to background subtraction but were smoothed using a 2 × 30-m/z window and Savitzky-Golay algorithm.
Electronic supplemental material
The supplemental material includes two figures. Figure 10 shows ESI mass spectra acquired under optimized instrumental conditions of the his6Tus–TerB complex (10 μM) in 800, 1400, and 2000 mM NH4OAc at pH 8.0 showing the m/z range 1500–4500. There is a decrease in the abundance of ions from the complex with increasing NH4OAc concentration concomitant with an increase in abundance of ions from free his6Tus and DNA. Figure 11 shows a comparison of the relative ESI-MS response factors of free Tus and a Tus–TerB complex. A plot of Tusfree/Tustotal and Tuscomplex/Tustotal determined by comparing the relative intensities of the relevant ions in the ESI mass spectrum against the relative amounts of free and complexed Tus added to the solution. In this experiment, the his6Tus–TerB complex was prepared and titrated with A173T in 800 mM NH4OAc at pH 8.0. ESI mass spectra were acquired of mixtures in which Tustotal was maintained at 10 μM. The following relative amounts of A173T and his6Tus–TerB, respectively, were used in this experiment: 0 and 10 μM, 2 μM and 8 μM, 4 μM and 6 μM, 6 μM and 4 μM, 8 μM and 2 μM. This experiment compares the relative ESI-MS response factors of free (A173T) and complexed Tus (his6Tus–TerB).
Acknowledgments
The Australian Research Council (ARC) supported this work. Grants from the ARC and the University of Wollongong enabling the purchase of the electrospray mass spectrometer are gratefully acknowledged. We thank Dr. Cameron Neylon for providing some of the protein samples.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1101/ps.27702.
References
- Cech, N.B. and Enke, C.G. 2000. Relating electrospray ionization response to nonpolar character of small peptides. Anal. Chem. 72 2717–2723. [DOI] [PubMed] [Google Scholar]
- Cheng, X., Harms, A.C., Goudreau, P.N., Terwilliger, T.C., and Smith, R.D. 1996a. Direct measurement of oligonucleotide binding stoichiometry of gene V protein by mass spectrometry. Proc. Natl. Acad. Sci. 93 7022–7027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, X., Morin, P.E., Harms, A.C., Bruce, J.E., Ben-David, Y., and Smith, R.D. 1996b. Mass spectrometric characterization of sequence-specific complexes of DNA and transcription factor PU.1 DNA binding domain. Anal. Biochem. 239 35–40. [DOI] [PubMed] [Google Scholar]
- Constantopoulos, T.L., Jackson, G.S., and Enke, C.G. 1999. Effects of salt concentration on analyte response using electrospray ionization mass spectrometry. J. Am. Soc. Mass Spectrom. 10 625–634. [DOI] [PubMed] [Google Scholar]
- Coskun-Ari, F.F. and Hill, T.M. 1997. Sequence-specific interactions in the Tus-Ter complex and the effect of base pair substitutions on arrest of DNA replication in Escherichia coli. J. Biol. Chem. 272 26448–26456. [DOI] [PubMed] [Google Scholar]
- Coskun-Ari, F.F., Skokotas, A., Moe, G.R., and Hill, T.M. 1994. Biophysical characteristics of Tus, the replication arrest protein of Escherichia coli. J. Biol. Chem. 269 4027–4034. [PubMed] [Google Scholar]
- Craig, T.A., Benson, L.M., Tomlinson, A.J., Veenstra, T.D., Naylor, S., and Kumar, R. 1999. Analysis of transcription complexes and effects of ligands by microelectrospray ionization mass spectrometry. Nat. Biotechnol. 17 1214–1218. [DOI] [PubMed] [Google Scholar]
- Gottlieb, P.A., Wu, S., Zhang, X., Tecklenburg, M., Kuempel, P., and Hill, T.M. 1992. Equilibrium, kinetic, and footprinting studies of the Tus-Ter protein-DNA interaction. J. Biol. Chem. 267 7434–7443. [PubMed] [Google Scholar]
- Griffiths, W.J., Jonsson, A.P., Liu, S., Rai, D.K, and Wang, Y. 2001. Electrospray and tandem mass spectrometry in biochemistry. Biochem. J. 355 545–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha, J.H., Capp, M.W., Hohenwalter, M.D., Baskerville, M., and Record, M.T. 1992. Thermodynamic stoichiometries of participation of water, cations and anions in specific and non-specific binding of lac repressor to DNA. Possible thermodynamic origins of the "glutamate effect" on protein-DNA interactions. J. Mol. Biol. 228 252–264. [DOI] [PubMed] [Google Scholar]
- Hagmar, P., Bailey, M., Tong, G., Haralambidis, J., Sawyer, W.H., and Davidson, B.E. 1995. Synthesis and characterization of fluorescent oligonucleotides. Effect of internal labeling on protein recognition. Biochim. Biophys. Acta 1244 259–268. [DOI] [PubMed] [Google Scholar]
- Hofstadler, S.A. and Griffey, R.H. 2001. Analysis of noncovalent complexes of DNA and RNA by mass spectrometry. Chem. Rev. 101 377–390. [DOI] [PubMed] [Google Scholar]
- Jarrold, M.F. 1999. Unfolding, refolding, and hydration of proteins in the gas phase. Acc. Chem. Res. 32 360–367. [Google Scholar]
- Kamada, K., Horiuchi, T., Ohsumi, K., Shimamoto, N., and Morikawa, K. 1996. Structure of a replication-terminator protein complexed with DNA. Nature 383 598–603. [DOI] [PubMed] [Google Scholar]
- Kapur, A., Beck, J.L., and Sheil, M.M. 1999. Observation of daunomycin and nogalamycin complexes with duplex DNA using electrospray ionization mass spectrometry. Rapid Commun. Mass Spectrom. 13 2489–2497. [DOI] [PubMed] [Google Scholar]
- Konermann, L. and Douglas, D.J. 1998. Unfolding of proteins monitored by electrospray ionization mass spectrometry: A comparison of positive and negative ion modes. J. Am. Soc. Mass Spectrom. 9 1248–1254. [DOI] [PubMed] [Google Scholar]
- Lim, H.-K., Hsieh, Y.L., Ganem, B., and Henion, J. 1995. Recognition of cell wall peptide ligands by vancomycin group antibiotics: Studies using ion spray mass spectrometry. J. Mass Spectrom. 30 708–714. [Google Scholar]
- Loo, J.A. 1997. Studying noncovalent protein complexes by electrospray ionization mass spectrometry. Mass Spectrom Rev. 16 1–23. [DOI] [PubMed] [Google Scholar]
- Neylon, C., Brown, S.E., Kralicek, A.V., Miles, C.S., Love, C.A., and Dixon, N.E. 2000. Interaction of the Escherichia coli replication terminator protein (Tus) with DNA: A model derived from DNA-binding studies of mutant proteins by surface plasmon resonance. Biochemistry 39 11989–11999. [DOI] [PubMed] [Google Scholar]
- Potier, N., Donald, L.J., Chernushevich, I., Ayed, A., Ens, W., Arrowsmith, C.H, Standing, K.G., and Duckworth, H.W. 1998. Study of a noncovalent trp repressor:DNA operator complex by electrospray ionization time-of-flight mass spectrometry. Protein Sci. 7 1388–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. 1989. Molecular cloning: A laboratory manual, 2d ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Sannes-Lowery, K.A., Griffey, R.H., and Hofstadler, S.A. 2000. Measuring dissociation constants of RNA and aminoglycoside antibiotics by electrospray ionization mass spectrometry. Anal. Biochem. 280 264–271. [DOI] [PubMed] [Google Scholar]
- Schwabe, J.W.R. 1997. The role of water in protein-DNA interactions. Curr. Opin. Struct. Biol. 7 126–134. [DOI] [PubMed] [Google Scholar]
- Schwartz, B.L., Bruce, J.E., Anderson, G.A., Hofstadler, S.A., Rockwood, A.L., Smith, R.D., Chilkoti, A., and Stayton, P.S. 1995. Dissociation of tetrameric ions of non-covalent streptavidin complexes formed by electrospray ionization. J. Am. Soc. Mass Spectrom. 6 459–465. [DOI] [PubMed] [Google Scholar]
- Skokotas, A., Wrobleski, M., and Hill, T.M. 1994. Isolation and characterization of mutants of tus, the replication arrest protein of Escherichia coli. J. Biol. Chem. 269 20446–20455. [PubMed] [Google Scholar]
- Wan, K.X., Gross, M.L., and Shibue, T. 2000. Gas-phase stability of double-stranded oligodeoxynucleotides and their noncovalent complexes with DNA-binding drugs as revealed by collisional activation in an ion trap. J. Am. Soc. Mass Spectrom. 11 450–457. [DOI] [PubMed] [Google Scholar]
- Wang, G. and Cole, R.B. 1994. Disparity between solution-phase equilibria and charge state distributions in positive-ion electrospray mass spectrometry. Org. Mass Spectrom. 29 419–427. [Google Scholar]
- Wickham, G., Iannitti, P., Boschenok, J., and Sheil, M.M. 1995. Electrospray ionization mass spectrometry of covalent ligand-oligonucleotide adducts: Evidence for specific duplex ion formation. J. Mass Spectrom. 30 S197–S203. [PubMed] [Google Scholar]
- Xu, N., Pasa-Tolic, L., Smith, R.D., Ni, S., and Thrall, B.D. 1999. Electrospray ionization-mass spectrometry study of the interaction of cisplatin-adducted oligonucleotides with human XPA minimal binding domain protein. Anal. Biochem. 272 26–33. [DOI] [PubMed] [Google Scholar]