Abstract
The replacement of Phe120 with other hydrophobic residues causes a decrease in the activity and thermal stability in ribonuclease A (RNase A). To explain this, the crystal structures of wild-type RNase A and three mutants—F120A, F120G, and F120W—were analyzed up to a 1.4 Å resolution. Although the overall backbone structures of all mutant samples were nearly the same as that of wild-type RNase A, except for the C-terminal region of F120G with a high B-factor, two local conformational changes were observed at His119 in the mutants. First, His119 of the wild-type and F120W RNase A adopted an A position, whereas those of F120A and F120G adopted a B position, but the static crystallographic position did not reflect either the efficiency of transphosphorylation or the hydrolysis reaction. Second, His119 imidazole rings of all mutant enzymes were deviated from that of wild-type RNase A, and those of F120W and F120G appeared to be "inside out" compared with that of wild-type RNase A. Only ∼1 Å change in the distance between Nɛ2 of His12 and Nδ1 of His119 causes a drastic decrease in kcat, indicating that the active site requires the strict positioning of the catalytic residues. A good correlation between the change in total accessible surface area of the pockets on the surface of the mutant enzymes and enthalpy change in their thermal denaturation also indicates that the effects caused by the replacements are not localized but extend to remote regions of the protein molecule.
Keywords: Crystal structure, ribonuclease A, active site, His119, thermal stability, accessible surface area
Bovine pancreatic ribonuclease (RNase A; EC 3.1.27.5; Cuchillo et al. 1997; Raines 1998) is an endonuclease that cleaves at the 3`-end of pyrimidine nucleosides. His12, His119, and Lys41 comprise the catalytic site, and several other amino acid residues serve as substrate binding subsites (i.e., Thr45, Asp83, and Phe120 in B1 subsite; Gln11 in P1 subsite; and Asn71 and Glu111 in B2 subsite). Phe120, which not only occupies a part of the B1 binding subsite (delCardayre and Raines 1994) but also is located close to the active site, plays an important role in the construction of the active site for efficient catalysis (Hayashi et al. 1973; Tanimizu et al. 1998; Chatani et al. 2001). We postulated that Phe120 plays a role in retaining the unique properties of His12 and His119 as well as some other functionally important amino acid residues, such as Asp121 (Cederholm et al. 1991; Quirk et al. 1998; Schultz et al. 1998b), Lys7, Arg10, and Lys66 (Boix et al. 1994; Fisher et al. 1998), and the disulfide bridges Cys65–Cys72 and Cys40–Cys95 (Klink et al. 2000). In our previous study, it was shown that the mutagenic replacement of Phe120 had no effect on the pKa of either His12 or His119 but caused a significant decrease in the rate of carboxymethylation by bromoacetate and iodoacetate at pH 5.5 with a concomitant decrease in activity, indicating that the mutagenic replacement of Phe120 affects the spatial positions of His12 and/or His119 (Chatani et al. 2001). At the same time, Phe120 also appeared to be important in maintaining a stable protein structure.
In this study, to investigate if the change in spatial positions of the two histidine residues is in fact caused by the replacement of Phe120 and if a conformational change does occur in the Phe120 mutant enzymes, the degree of the conformational changes—detailed conformational changes with respect to His12 and His119—as the result of mutagenic replacement of Phe120 were evaluated via an analysis of the crystal structures of F120A, F120G, and F120W mutant RNase A by X-ray crystallography at high resolution. Because a subtle conformational change could have an effect on catalytic activity, as has been previously suggested, wild-type RNase A was also crystallized under the same condition as other mutant enzymes, and its structure was used for a precise comparison with the mutant RNase A samples.
Results
Crystallization, data collection, and structural refinement
The crystals of wild-type, F120A, F120G, and F120W RNase A appeared within a day and grew to a size of ∼150 × 150 × 150 μm. We also attempted to crystallize F120L mutant RNase A, the activity, thermal stability, and carboxymethylation rate of which were also analyzed in our previous study (Chatani et al. 2001), but only a cluster of fine needle crystals was obtained in the similar condition as that of the wild-type RNase A. A single crystal was once obtained, but this was not reproducible and we could not obtain enough data.
The crystals of the wild-type and all mutant RNase A (F120A, F120G, and F120W RNase A) belonged to space group P3221, with similar values of unit cell dimensions (Table 1). The highest resolution of data obtained in this study was 1.35 Å for the wild-type enzyme, 1.0 Å for the F120A and F120W enzymes, and 1.2 Å for the F120G enzyme. The statistics of the data collection were judged to be fit for use as listed in Table 1.
Table 1.
Statistics of data collection and structure refinement
| Enzyme | Wild type | F120A | F120G | F120W |
| Data collection | ||||
| Space group | P3221 | |||
| Unit cell dimension (Å) | a = b = 64.05, c = 63.36 | a = b = 63.66, c = 63.49 | a = b = 63.73, c = 63.49 | a = b = 63.90, c = 63.33 |
| (deg) | γ = 120 | γ = 120 | γ = 120 | γ = 120 |
| X-ray source | SPring-8 BL40B2 | SPring-8 BL44B2 | SPring-8 BL40B2 | SPring-8 BL44B2 |
| Wavelength (Å) | 0.968641 | 0.7 | 0.7 | 0.7 |
| Detector | Rigaku RAXIS IV++ IP | ADSC Quantum 4R CCD | Rigaku RAXIS IV ++ IP | ADSC Quantium 4D CCD |
| Mount | Cryo protectant (300 mg/ml trehalose) | |||
| Maximum resolution (Å) | 1.35 | 1.00 | 1.20 | 1.00 |
| Data collection resolution (Å) | 55.5–1.35 | 16.8–1.00 | 41.7–1.20 | 16.8–1.00 |
| Number of total reflections | 241,908 | 1,446,353 | 404,905 | 886,320 |
| Number of unique reflections | 33,471 | 80,089 | 41,727 | 80,099 |
| Completeness (%) | 98.8 | 99.7 | 88.8 | 99.6 |
| Average I/σ | 14.3 | 36.9 | 12.6 | 34.9 |
| Rmerge | 0.05 | 0.07 | 0.04 | 0.06 |
| Structure refinement | ||||
| Refinement resolution (Å) | 6.00–1.40 | 5.00–1.40 | 6.00–1.40 | 6.00–1.40 |
| R-factor | 0.204 | 0.201 | 0.210 | 0.218 |
| R-free | 0.242 | 0.254 | 0.265 | 0.272 |
| RMS (bond distances) (Å) | 1.2 | 1.2 | 1.1 | 1.2 |
| RMS (bond angle) (deg) | 0.006 | 0.005 | 0.005 | 0.005 |
The structures were refined by rigid-body, positional, slow-cool, and b-refinement in X-PLOR. At a refinement of up to the 2.0-Å resolution, ∼30 water molecules were picked up and followed by continuous structural refinement. Ultimately, 115 to 238 water molecules were picked up. The fitness of electron density was evaluated with a model that was constructed at each step during the refinement. Manual corrections were performed by checking the 2Fo − Fc map and Fo − Fc map. All structures were refined up to a maximum resolution of 1.4 Å. The R-factor was 20% to 22% in all enzymes (Table 1). Differences between Rfree and Rall were <5.5% in all cases. The electron density at position 120 in the F120A, F120G, and F120W mutant RNase A indicated that the mutagenic alteration of Phe120 to alanine, glycine, and tryptophan, respectively, had been accomplished. The completed coordinates of the wild-type, F120A, F120G, and F120W RNase A have been deposited to the Protein Data Bank (PDB) with the PDB codes 1FS3, 1EIC, 1EID, and 1EIE, respectively.
Overall backbone structures
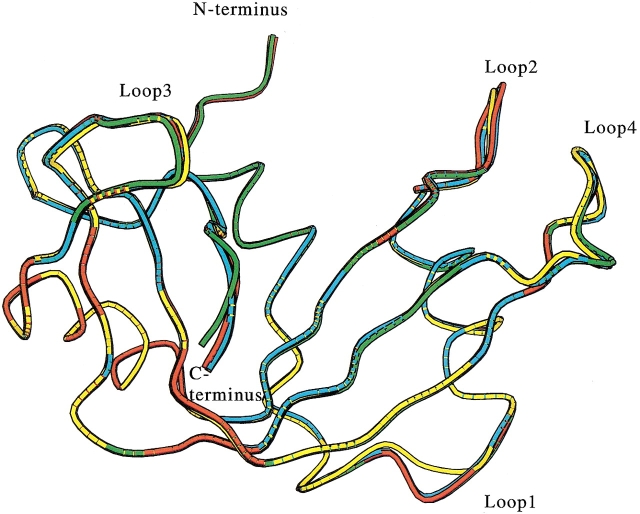
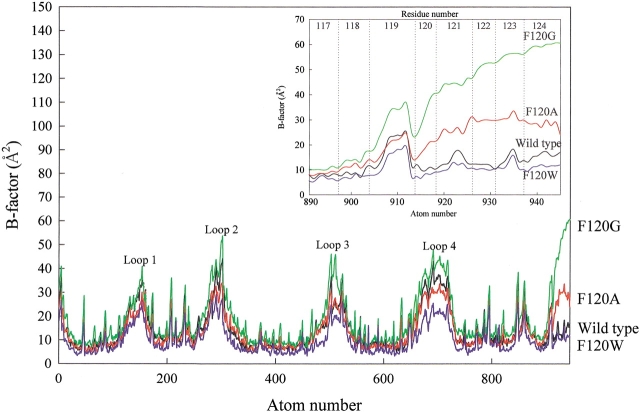
The overall backbone structures of F120A and F120W RNase A were very similar to that of wild-type RNase A, except for the loop regions (Fig. 1 ▶), where the B-factors were higher than that of other regions of the wild-type and mutant RNase A (Fig. 2 ▶), indicating flexibility in the loop regions. The backbone structures of the C-terminal regions (residues 120 to 124) of F120A and F120W were well superimposed, but the backbone structure of the F120G RNase A deviated from that of the wild-type RNase A, despite good superimposition of the residual structure. The B-factors of the C-terminal region in F120G RNase A increased by ∼20 Å2 relative to that in wild-type RNase A, whereas those in F120A RNase A were only slightly larger than that in wild-type RNase A (Fig. 2 ▶).
Fig. 1.
Superimposition of the backbone structures of wild-type and mutant RNase A. This figure was drawn with the MOLSCRIPT program (Kraulis 1991). Yellow, red, green, and cyan are wild-type, F120A, F120G, and F120W enzymes, respectively. Loops 1, 2, 3, and 4 represent the loop regions of 15–25, 34–41, 65–72, and 87–97, respectively.
Fig. 2.
B-factor of wild-type and mutant RNase A. Loops 1, 2, 3, and 4 correspond to those in Fig. 1 ▶. Atoms of the side-chain at 120 are omitted from the plots. (Inset) An enlargement of the C-terminal regions. Wild-type, F120A, F120G, and F120W RNase A are drawn with black, red, green, and blue, respectively.
Substrate binding sites
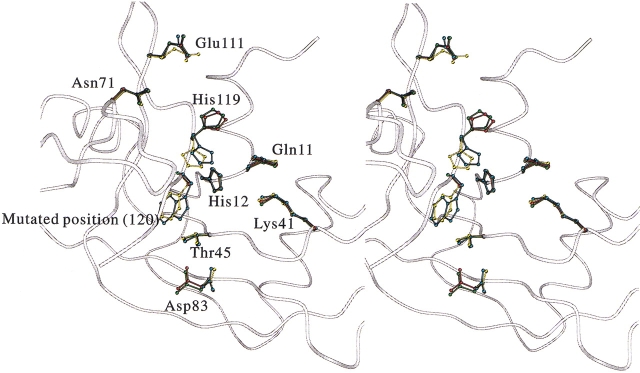
Amino acid residues comprising the B1, B2, and P1 substrate binding subsites revealed no significant conformational changes by the replacement of Phe120 with alanine, glycine, and tryptophan, as shown in Figure 3 ▶. Asp83 and Glu111 were not well superimposed, but these positional changes were not caused by the replacement of Phe120 because their positions are sensitively affected by crystallization conditions as seen by the coordinates of RNase A deposited to the PDB. The spatial position of amino acid residues at the position of 120 showed only slight differences among the wild-type and mutant enzymes as judged by root-mean-square deviation (RMSD) values: The RMSD values of the Cα carbon at the position of 120 between wild-type RNase A and F120A, F120G, or F120W RNase A were 0.21 Å, 0.43 Å, and 0.30 Å, respectively. The RMSD values of the Cβ carbons between wild-type RNase A and F120A or F120W enzymes were 0.28 Å and 0.57 Å, respectively. The benzene ring of Phe120 in the wild-type RNase A and the indole ring of Trp120 in F120W RNase A also orientated very similarly (χ1 and χ2 of His119 in the wild-type RNase A were 173° and −85°, respectively, and those in F120W RNase A were −168° and −90°, respectively). The B-factors of Trp120 in F120W RNase A and Phe120 in the wild-type RNase A remained the same. However, the B-factors increased as Phe120 was substituted with smaller hydrophobic residues, in the order of phenylalanine, alanine, and glycine (Fig. 2 ▶).
Fig. 3.
Stereo view of the superimposed structures of the active site in the wild-type and mutant RNase A. This figure was drawn with the MOLSCRIPT program (Kraulis 1991). Backbone structure of wild-type RNase A is drawn in the form of a white tube. Ball-and-stick models show the coordinates of Gln11, His12, Lys41, Thr45, Asn71, Asp83, Glu111, His119, and amino acid residue at the position 120. Yellow, red, green, and cyan are wild-type, F120A, F120G, and F120W RNase A, respectively.
Catalytic residues, His12, Lys41, and His119
Positional changes in the catalytic residues of His12 and Lys41 were less than those observations among wild-type and mutant RNase A (Fig. 3 ▶), and their B-factors were also similar (Fig. 2 ▶).
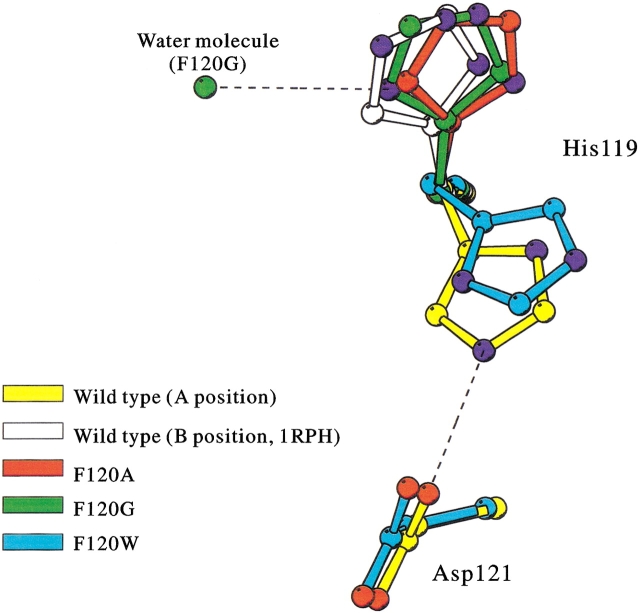
The main-chains of His119 in the wild-type and Phe120 mutant RNase A coincided well, whereas side-chains of His119 in mutant RNase A deviated from the original positions, as shown in Figure 3 ▶. In addition, although His119 in the wild-type and F120W RNase A were in the A position and directed toward position 120, His119 in F120A and F120G RNase A were in the B position and directed toward position 118 (Fig. 4 ▶; see Discussion for A and B positions of His119).
Fig. 4.
Superimposition of His119 side-chains of wild-type and mutant RNase A. Yellow, red, green, cyan, and white are wild-type (A position), F120A (B position), F120G (B position), F120W (A position), and wild-type (B position; PDB code 1RPH) enzymes, respectively. Nitrogen and oxygen atoms in His119 and Asp121 are colored by blue and red, respectively. A water molecule observed near the His119 of F120G RNase A is also shown in green. Dashed lines show hydrogen bonds connected with the His119 side-chains.
The position of His119 in F120W RNase A, which assumes the A position, was compared with that in wild-type RNase A, which also assumes the A position: The ϕ and ψ angles of His119 in the wild-type (141° and −158°, respectively) and F120W RNase A (140° and −156°, respectively) were very similar, but the χ1 and χ2 angles of His119 in wild-type (156° and 87°, respectively) and F120W RNase A (−177° and −118°, respectively) were clearly different. It is interesting to note that the face of the imidazole ring in F120W RNase A appeared to turn inside out in the present model (Fig. 4 ▶), although this conclusion is tentative because it is impossible to distinguish N from C atoms of the imidazole ring by the present electron density map. When the imidazole ring of His119 in F120W RNase A was rotated by 180° from the original face in a manual operation using computer graphics, followed by energy minimization, the ring failed revert to the orientation that existed before the manual operation. On the other hand, an original hydrogen bond between Nɛ2 of His119 and Oδ1 of Asp121, which is important in orientating the proper tautomeric form of His119 (Cederholm et al. 1991; Quirk et al. 1998; Schultz et al. 1998b), was lost in F120W even if the imidazole ring of F120W was rotated by 180° (Fig. 4 ▶), supporting the validity of our present proposed structure.
The position of His119 in F120A and F120G RNase A, which are in the B position, were compared with that of wild-type RNase A in B position determined by Zegers et al. (1994; PDB identification code 1RPH). The ϕ and ψ angles of His119 (in the B position) in the wild-type (134° and −152°, respectively), F120A (146° and −155°, respectively), and F120G RNase A (151° and −151°, respectively) were all similar. However, the χ1 and χ2 angles of His119 for the wild-type (−54° and −51°, respectively), F120A (−72° and −45°, respectively), and the F120G RNase A (−66° and 126°, respectively) were different from each other. The His119 imidazole ring of F120G RNase A also turned inside out as did that of F120W (Fig. 4 ▶). Opposed to the case of F120W RNase A, however, this inside-out orientation was preferred to the original orientation of the wild-type RNase A, probably because of a hydrogen bond between Nδ1 of His119 and a water molecule (Fig. 4 ▶). The B-factor of His119 in F120G RNase A was higher than that of wild-type and F120A RNase A.
Cavities or pockets
Cavity and pocket sizes of wild-type and three mutant enzymes, and RNase A in which His119 occupies B position (1RPH), were analyzed by CASTp program (Liang et al. 1998) with a solvent probe of 1.4 Å. As a result, although no cavity was detected, 8 to 12 pockets were found in all crystal structures, as shown in Table 2. The pockets were dispersed all over the protein surface. Phe120 of wild-type (1FS3), Phe120 of wild-type (1RPH; Zegers et al. 1994), Ala120 of F120A, Gly120 of F120G, and Trp120 of F120W belonged to the pockets 7, 10, 8, 9, and 11, respectively (Table 2).
Table 2.
Accessible surface area of pockets detected by CASTp programa
| Pocket no. | Wild type (His119 takes A position) | Wild type (His119 takes B position)b | F120A | F120G | F120W |
| 1 | 6.835 | 0.064 | 0.184 | 0.006 | 0.181 |
| 2 | 10.823 | 0.013 | 0.017 | 0.011 | 0.022 |
| 3 | 10.195 | 0.030 | 4.442 | 1.581 | 1.178 |
| 4 | 4.546 | 0.028 | 14.260 | 13.120 | 0.102 |
| 5 | 26.683 | 0.934 | 19.791 | 25.332 | 1.082 |
| 6 | 21.007 | 2.466 | 45.918 | 23.093 | 10.120 |
| 7 | 71.228c | 16.940 | 28.422 | 50.264 | 9.290 |
| 8 | 113.146 | 23.072 | 173.518c | 94.850 | 8.063 |
| 9 | — | 17.275 | — | 164.66c | 18.312 |
| 10 | — | 55.399c | — | — | 21.601 |
| 11 | — | — | — | — | 39.737c |
| 12 | — | — | — | — | 96.462 |
| Total accessible surface area (ASAtotal) | 264.463 | 116.221 | 286.552 | 372.917 | 206.15 |
| ΔASAtotald | 0 | 0 | 170.331 | 256.696 | −58.313 |
a Unit in Å2.
b Calculated from the crystal structure of PDB code 1RPH (Zegers et al. 1994) from which His119 at A position was deleted.
c Accessible surface area of the pocket to which side chain at the position 120 belongs.
d ΔASAtotal = ASAtotalmutant − ASAtotalwild type. The value 264.463 was used as ASAtotal wild type in the case of F120W in which His119 takes A position, and 116.221 in the case of F120A and F120G in which His119 takes B position.
Discussion
Even subtle conformational changes in the three Phe120 mutant RNase A were detected by the crystallographic structural analysis up to 1.4-Å resolution with reasonable statistical values. The mutagenic replacement of Phe120 caused a positional change in His119, whereas the B1, B2, and P1 substrate binding subsites remained the same. This finding is consistent with our previous results based on kinetic studies on cytidilyl-3`,5`-adenosine (CpA) and cytidine-2`,3`-cyclic monophosphate (C>p) and inhibition by phosphate anion. Unexpectedly, the mutagenic alternation of Phe120 results in no change in the spatial position of His12, even though the benzene ring of Phe120 and the imidazole ring of His12 face each other within a distance of 4.5 Å, which is sufficiently short to make a cation-aromatic interaction possible (Rico et al. 1986; Shoemaker et al. 1990; Loewenthal et al. 1992). This fact is consistent with the previous report that the mutagenic replacement of Phe120 does not lead to a change in the pKa of His12. It had been suggested that Phe120 stabilizes the transition state of the RNase A reaction by π-electron stacking between Phe120 and substrates (Zegers et al. 1994; Tanimizu et al. 1998). However in the present case, the kcat value of F120W mutant RNase A—in which the indole ring of Trp120 is orientated similarly with the benzene ring of Phe120 in wild-type RNase A, and therefore π-electron stacking between the indole ring of Trp120 and the substrate is possibly made—decreases, without supporting this suggestion. Under these considerations, it can be concluded that the positional change of His119 induced by the mutagenic substitution of Phe120 is a major cause in decreasing the activity.
The positional change of His119 by mutagenic replacement of Phe120
It has been suggested that His119 position of RNase A fluctuates between the A and B positions in the aqueous solution. In the crystal state, the location of His119 is visible in A and/or B positions, depending on the crystallization conditions (Borkakoti et al. 1982; Wlodawer et al. 1982; Martin et al. 1987). Although the wild-type and mutant RNase A were crystallized under similar conditions, in which it would be expected that His119 of all enzymes assumed the A position, the His119 imidazole rings of wild-type and F120W RNase A assumed the A position; those of F120A and F120G RNase A, the B position. A hydrogen bond between His119 and Asp121 in the wild-type enzyme (de Mel et al. 1992) is not essential in favoring the A position because His119 in the F120W mutant does not contain this hydrogen bond but still assumes the A position. An increase in salt concentration or a decrease in pH favors the B position of His119 in the wild-type RNase A (de Mel et al. 1994b; Fedorov et al. 1996), indicating that a possible ionic interaction between the π-electron of Phe120 and the imidazole ring of His119 makes the A position of His119 preferable. Such ionic interaction could be present between the π electrons of the indole ring of Trp120 in F120W but not in the F120A and F120G enzymes, which favor the B position over the A position in the crystal. However, this is not the case for an aqueous solution, in which His119 of all the Phe120 enzymes would fluctuate between the A and B positions because the transition energy is very low (de Mel et al. 1994b). Thus, in the substrate-free state, the location of His119 at the A or B position is a crystallographic artifact and can be ignored in the case of an aqueous solution.
When wild-type RNase A is complexed with d(CpA) and 3`-CMP, the His119 assumes the A and B positions, respectively (Zegers et al. 1994), indicating that the His119 position changes during the reaction process, that is, the A position during transphosphorylation and the B position for the case of the hydrolysis reaction (Borkakoti 1983). However, this is inconsistent with the present result because kcat of the F120W and F120A mutant RNase A was affected more in the hydrolysis reaction than in the transphosphorylation, whereas the kcat for the F120G enzyme was more affected in the transphosphorylation reaction than in the hydrolysis reaction (Chatani et al. 2001). These results show that the preference of the His119 positions is not reflected in the total enzyme activity. It is reasonable to conclude that His119 exists in one position during the two continuous reactions, consistent with a recent proposal made by Schultz et al. (1998b).
The possibility has been suggested that the face of His119 in F120W RNase A is rotated by 180° from that of wild-type enzyme, which is probably caused by the loss of a hydrogen bond between Asp121 and His119. The hydrogen bond is thus important in preventing the rotation of the imidazole ring but is not significant in maintaining the proper pKa of His119 because the pKa of His119 in F120W RNase A was at the same level (Tanimizu et al. 1998). But the flipping of His119 should be confirmed with an additional experiment at higher resolution or by nuclear magnetic resonance study for concluding the role of the hydrogen bond between Asp121 and His119.
The face of His119 (B position) in the F120G RNase A crystal is also rotated by 180°. When the crystal structure of the F120G enzyme in which His119 assumes the A position is determined, this will clarify the issue of whether the rotation is also caused by the loss of the hydrogen bond. However, when the side-chain of His119 is rotated around χ1 and χ2 to dihedral angles of ∼159° and −137°, respectively, which are the rotation angles from the B position to the A position in the case of wild-type RNase A, and the imidazole ring then turns inside out (180° rotation), no hydrogen bond was formed between Asp121 and His119. Therefore, the rotation of His119 in F120G is possibly caused by the loss of the hydrogen bond. Even if Nɛ2 of His119 in the A position and the Oδ1 of Asp121 of F120G RNase A is sufficiently close for forming a hydrogen bond, the high flexibility of His119 in F120G RNase A (see B-factor in Fig. 2 ▶) prevents the formation of a hydrogen bond between His119 and Asp121.
Relationship between the distance from Nɛ2 of His12 to Nδ1 of His119 and activity
It has been known that the two nitrogen atoms Nɛ2 of His12 and Nδ1 of His119 participate in catalysis as an acid and base (Cuchillo et al. 1997; Raines 1998). We previously estimated that positional changes in His12 and/or His119 lower activity in Phe120 mutant enzymes from our results of carboxymethylation experiments, and in this study, we found a correlation between the deviation in distance from Nɛ2 of His12 and Nδ1 of His119 in the free form and enzyme activity (Table 3): As the absolute deviation increases, the activity decreases. Here, it should be noted that the highest deviation was only 1.3 Å (F120G), which results in a decrease in 93.3% of kcat for CpA. Although the scale of the deviation was much smaller than expected, the similar correlation is also observed in the other sets of mutant enzymes, that is, D121N and D121A and in semisynthetically prepared F120Y and F120L RNase A (it should be noted that the pKa of His12 and 119 of these mutant enzymes are the same as that of the wild-type enzyme; de Mel et al. 1994a; Schultz et al. 1998b), all of the deviations of which were <1.0 Å (Table 3). The obvious correlations shown in Table 3 indicate the possibility that a 1-Å deviation might drastically reduce activity. This conclusion is very surprising, because the flexibility of the active site of enzymes, the so-called induced fit, has been known to the active site of RNase A from the nuclear magnetic resonance and X-ray crystallographic studies, in which the active site conformation changed by substrate binding (Arus et al. 1982), and the temperature factors were lowered by the low temperature, accompanied by the loss of activity (Rasmussen et al. 1992). Even if a 1-Å deviation could be diminished by the induced fit, the obvious correlation between the deviation in distance from Nɛ2 of His12 and Nδ1 of His119 in the free form of enzymes and enzymatic activity indicates that the diminished deviation might still drastically reduce activity, namely, such conformational flexibility does not cover the 1-Å deviation to result in a dramatic decrease in activity. The active site of RNase A seems to involve two contrary features at the same time: flexibility and strictness. Attempts to determine the crystal structures of complexes with substrate analogs are underway to analyze the degree of the deviation after the induced fit to accept the substrate.
Table 3.
Distance between Nɛ2 of His12 and Nδ1 of His119, and kcat in wild-type and mutant RNase A
| Enzyme | Position of His119 | Distanced (Å) | Substrate | kcat (%) | Reference |
| Wild typea | A | 6.4 | CpA, C>p, UpA, U>p | 100 | |
| F120Wa | A | 7.5 (+1.1) | CpA | 26e | |
| C>p | 8.5e | ||||
| Wild type (1RPH)b | B | 7.4 | CpA, C>p | 100 | Zegers et al. 1994 |
| F120Ab | B | 6.6 (−0.8) | CpA | 72e | |
| C>p | 23e | ||||
| F120Gb | B | 8.7 (+1.3) | CpA | 6.7e | |
| C>p | 14.4e | ||||
| D121N (3RSD)a | A | 6.3 (−0.1) | UpA | 21 | Schultz et al. 1998b |
| U>p | 42 | ||||
| D121A (4RSD)a | A | 5.8 (−0.6) | UpA | 3.4 | Schultz et al. 1998b |
| U>p | 5.1 | ||||
| Wild type (1-118:111-124)(1SRN)c | B | 7.3 | C>p | 100 | Martin et al. 1987 |
| F120Y (1-118:111-124)(1SSB)c | B | 6.9 (−0.4) | C>p | 85 | de Mel et al. 1994a |
| F120L (1-118:111-124)(1SSA)c | B | 6.8 (−0.5) | C>p | 13 | de Mel et al. 1994a |
a The structures of F120W, D121N, and D121A enzymes were compared with that of wild-type RNase A determined in this study.
b The structures of F120A, and F120G enzymes were compared with that of 1RPH wild-type RNase A.
c The structures of F120Y, and F120L semisynthetic RNase A were compared with that of 1SRN wild-type semisynthetic RNase A.
d Distance between Nɛ2 of His12 and Nδ1 of His119. Value in parenthesis shows the difference in the distance from that of wild-type RNase A.
e kcat data were taken from our previous papers (Tanimizu et al., 1998; Chatani et al. 2001).
A decrease in RNase A activity had been ascribed to the conformational change of the 65–72 loop region, according to the experimental results used by semisynthetic F120L, D121N, and D121A RNase A (de Mel et al. 1992, 1994a). This also appears to be supported by the present results, in that the loop region of the three Phe120 mutant RNase A appears to move away from the active site. However, the degrees of this movement in F120A, F120G, and F120W (i.e., 0.40, 0.55, and 0.34 Å, respectively) show no correlations with a decrease in activity. Moreover, all of the mutant enzymes maintain a hydrogen bond of similar length between the main-chain N of Lys66 and Oδ2 of Asp121. Because loop regions are usually flexible, as evidenced by their high B-factor (Fig. 2 ▶), the observed conformational differences in the loop regions of the present mutant enzymes are within experimental error. Thus, it can be concluded that the decrease in activity in mutant RNase A at Phe120 can be ascribed not to conformational changes in the loop region but rather to the shift of His119. At pH 2.2, the loop region of RNase A moves away from its C terminus, accompanied by a significant disorder in the C-terminal region (Ratnaparkhi and Varadarajan 1999). This shows that the conformation of the loop and the C-terminal regions cooperatively changes as the result of the mutation at Phe120, located in the C terminus.
Relationship of B-factor at C-terminal region with activity and stability
A larger decrease in the activity of F120G RNase A than would have been expected may be owing to not only the positional change in His119 but also an increase in the B-factor of His119 (Fig. 2 ▶). An increase in flexibility lowers the probability of juxtaposing His119, leading to a decrease in activity.
The flexibility of the C-terminal residues also affects conformational stability, because the B-factor around the C-terminal region increases as Phe120 is replaced by smaller hydrophobic residues in the order of F120G > F120A > wild type > F120W. This is the same order with entropy values of thermal denaturation. It is therefore entirely possible that the entropy change is the result of a change in flexibility in the C-terminal region, which is, in turn, the result of the replacement of Phe120.
Correlation of accessible surface area with thermal stability
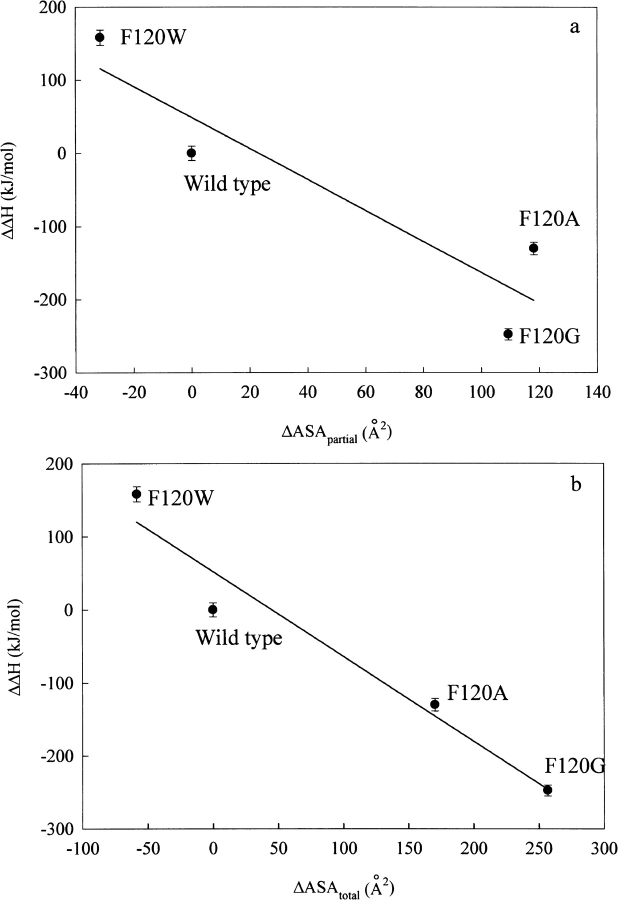
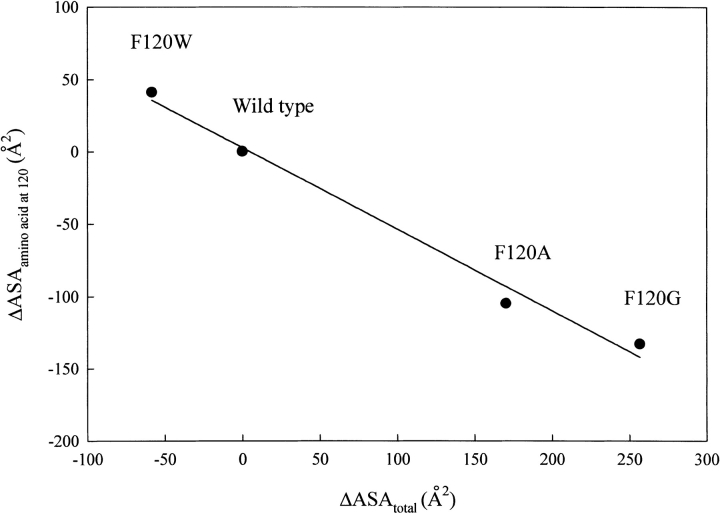
A linear correlation between the accessible surface area (ASA) of a cavity and thermal stability has been reported for some proteins (Eriksson et al. 1992; Takano et al. 1997; Coll et al. 1999). The Phe120 mutant RNase A used herein showed a linear correlation between the change in ASA of the pocket to which the amino acid residue at 120 belongs (ΔASApartial) and ΔΔH for thermal denaturation (Chatani et al. 2001), with a correlation coefficient of 0.92 (Fig. 5a ▶). A better correlation was obtained between the change in total ASA of all pockets (ΔASAtotal) and ΔΔH, with a correlation coefficient of 0.97 (Fig. 5b ▶). In addition, a good linear correlation exists between ΔASAtotal and the ASA of amino acid residue itself occupying the position 120 (ΔASAamino acid at 120; Miller et al. 1987), with a correlation coefficient of 0.99 (Fig. 6 ▶). These facts indicate that the change in volume at the position 120 as the result of the mutation affects not only the neighborhood of the substituted position but also the entire protein molecule, and these widespread conformational deviations cause the changes in enthalpy and entropy, thus destabilizing the protein structure. Phe120 appears to interact with a number of other amino acid residues as a part of the network of noncovalent bonds to build up the strict conformation of RNase A, which produces not only the maximum activity but also the optimum conformational stability of RNase A.
Fig. 5.
Correlation of ΔΔH in thermal denaturation with ΔASApartial (the change in the accessible surface area of the pocket to which amino acid residue at the position 120 belongs; a) and with ΔASAtotal (b). The lines were obtained by the least-squares method with correlation coefficients of 0.922 and 0.975, respectively.
Fig. 6.
Correlation of ΔASAtotal (change in the total accessible surface area; see Table 2) with ΔASAamino acid at 120, which was calculated by ASAsubstituting amino acid at 120 − ASAphenylalanine using the results by Miller et al. (1987) The line is obtained by the least-squares method with a correlation coefficient of 0.99.
In conclusion, the decrease in activity is caused by only a slight displacement and increase in the flexibility of His119. The entropy factor in the conformational stability is caused by an increase in flexibility in the C-terminal region. Thus, Phe120 is important in fixing the proper spatial position of His119 near the C-terminal region for efficient activity. The side-chain of Phe120 is in the backside of His119 and directs it toward a hydrophobic core. Thus, it is natural that Phe120 interacts not directly with His119 but with the hydrophobic core. The replacement of Phe120 with other amino acid residues would affect the hydrophobic core, thereby changing the position of His119. Such an indirect effect would also accompany the positional change of Asp121, and a hydrogen bond between His119 and Asp121 would be lost. Moreover, subtle conformational changes range over the entire protein molecule, thus causing a decrease in stability by the replacement of Phe120 and indicating the delicate nature of the protein conformation.
Materials and methods
Materials
F120A, F120G, and F120W mutant RNase A were produced and purified as previously described (Chatani et al. 2001). Commercial RNase A (type III-A, Sigma) was used as wild-type enzyme.
Crystallization
The purified enzyme solution was diluted with water 1000-fold and concentrated to 24 mg protein/mL. Wild-type and mutant RNase A were crystallized by the hanging-drop vapor-diffusion method according to Schultz et al. (1998a) with minor modifications: A 6-μL drop containing the protein solution and the reservoir solution in a ratio of 1:1 were equilibrated against 1.0 mL of the reservoir solution, which contained 100 mM sodium acetate buffer (pH 6.0), 30% to 35% (v/v) of saturated ammonium sulfate (35% for F120W RNase A, 34% for F120A RNase A, and 30% for wild-type and F120G RNase A), and 50% (v/v) of saturated NaCl by incubating at 25°C.
Data collection
X-ray diffraction data of F120W and F120A mutant RNase A were collected with an ADSC Quantum 4R CCD detector system at SPring-8 BL44B2 (Hyogo, Japan). X-ray diffraction data for the wild-type and F120G mutant RNase A were collected with a RIGAKU RAXIS IV++ imaging plate detector system at SPring-8 BL40B2 (Hyogo, Japan). During the data collection, the crystals were cooled to 100 K with a stream of nitrogen gas. Data from CCD detector system and the imaging plate detector system were processed and scaled using the MOSFLM program in Collaborative Computational Project No. 4 (1994) and the DENZO program (Otwinowski and Minor 1997), respectively. Details of the data collection are listed in Table 1.
Refinement of the crystal structure
The molecular structure was determined by the molecular replacement method using the X-PLOR program (Ver. 3.81; Brünger 1992). The coordinate of the PDB code 1RNM, which was stripped of all solvent molecules, was used as a starting model. The geometry of the main-chain and side-chains was analyzed using the PROCHECK program (Laskowski et al. 1993). The model was adjusted manually in the O program (Jones et al. 1991). After several cycles of least-square refinements and manual adjustments, water molecules were added to the model. In analyzing conformational change as the result of the mutagenic replacement of Phe120, the structures of wild-type and mutant RNase A were superimposed by use of backbone atoms using the Swiss-PdbViewer program (Guex and Peitsch 1997).
Acknowledgments
We thank Dr. Y. Katsube for advising us on the effective use of X-ray crystallography. We also thank Ms. K. Miura at BL40B2 and Dr. S. Adachi at BL44B2 beamline in SPring-8 for their help in data collection, and Ms. E. Yamashita for preparing the crystals of wild-type RNase A. This work was supported, in part, by a Grant-in-Aid for Scientific Research from the Ministry of Education, Science, Sports and Culture of Japan; a grant from Japan Society for the Promotion of Science (JSPS) for E.C.; and a grant from Japan Space Forum for H.M.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Abbreviations
ASA, accessible surface area
CpA, cytidilyl-3`,5`-adenosine
C>p, cytidine-2`,3`-cyclic monophosphate
RMSD, root-mean-square deviation
UpA, uridilyl-3`,5`-adenosine
U>p, uridine-2`,3`-cyclic monophosphate.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1101/ps.31102.
References
- Arus, C., Paolillo, L., Llorens, R., Napolitano, R., and Cuchillo, C.M. 1982. Evidence on the existence of a purine ligand-induced conformational change in the active site of bovine pancreatic ribonuclease A studied by proton nuclear magnetic resonance spectroscopy. Biochemistry 21 4290–4297. [DOI] [PubMed] [Google Scholar]
- Boix, E., Nogues, M.V., Schein, C.H., Benner, S.A., and Cuchillo, C.M. 1994. Reverse transphosphorylation by ribonuclease A needs an intact P2-binding site: Point mutations at Lys-7 and Arg-10 alter the catalytic properties of the enzyme. J. Biol. Chem. 269 2529–2534. [PubMed] [Google Scholar]
- Borkakoti, N. 1983. The active site of ribonuclease A from the crystallographic studies of ribonuclease-A-inhibitor complexes. Eur. J. Biochem. 132 89–94. [DOI] [PubMed] [Google Scholar]
- Borkakoti, N., Moss, D.S., and Palmer, R.A. 1982. Ribonuclease A: Least-squares refinement of the structure at 1.45 Å resolution. Acta Crystallogr. B 38 2210–2217. [Google Scholar]
- Brünger, A.T. 1992. X-PLOR version 3.1 Manual: A system for crystallography and NMR. Yale University, New Haven, CT.
- Cederholm, M.T., Stuckey, J.A., Doscher, M.S., and Lee, L. 1991. Histidine pKa shifts accompanying the inactivating Asp121-Asn substitution in a semisynthetic bovine pancreatic ribonuclease. Proc. Natl. Acad. Sci. 88 8116–8120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatani, E., Tanimizu, N., Ueno, H., and Hayashi, R. 2001. Structural and functional changes in bovine pancreatic ribonuclease A by the replacement of Phe120 with other hydrophobic residues. J. Biochem. 129 917–922. [DOI] [PubMed] [Google Scholar]
- Coll, M.G., Protasevich, I.I., Torrent, J., Ribo, M., Lobachov, V.M., Makarov, A.A., and Vilanova, M. 1999. Valine 108, a chain-folding initiation site-belonging residue, crucial for the ribonuclease A stability. Biochem. Biophys. Res. Commun. 265 356–360. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 (CCP4). 1994. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50 760–763. [DOI] [PubMed] [Google Scholar]
- Cuchillo, C.M., Vilanova, M., and Nogues, M.V. 1997. Pancreatic ribonucleases. In Ribonucleases: Structures and functions (eds. G. D'Alessio and J.F. Riordan), pp. 271–300, Academic Press, New York, NY.
- delCardayre, S.B. and Raines, R.T. 1994. Structural determinants of enzymatic processivity. Biochemistry 33 6031–6037. [DOI] [PubMed] [Google Scholar]
- de Mel, V.S., Martin, P.D., Doscher, M.S., and Edwards, B.F. 1992. Structural changes that accompany the reduced catalytic efficiency of two semisynthetic ribonuclease analogs. J. Biol. Chem. 267 247–256. [PubMed] [Google Scholar]
- de Mel, V.S., Doscher, M.S., Glinn, M.A., Martin, P.D., Ram, M.L., and Edwards, B.F. 1994a. Structural investigation of catalytically modified F120L and F120Y semisynthetic ribonucleases. Protein Sci. 3 39–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Mel, V.S., Doscher, M.S., Martin, P.D., and Edwards, B.F. 1994b. The occupancy of two distinct conformations by active-site histidine-119 in crystals of ribonuclease is modulated by pH. FEBS Lett. 349 155–160. [DOI] [PubMed] [Google Scholar]
- Eriksson, A.E., Baase, W.A., Zhang, X.J., Heinz, D.W., Blaber, M., Baldwin, E.P., and Matthews, B.W. 1992. Response of a protein structure to cavity-creating mutations and its relation to the hydrophobic effect. Science 255 178–183. [DOI] [PubMed] [Google Scholar]
- Fedorov, A.A., Joseph-McCarthy, D., Fedorov, E., Sirakova, D., Graf, I., and Almo, S.C. 1996. Ionic interactions in crystalline bovine pancreatic ribonuclease A. Biochemistry 35 15962–15979. [DOI] [PubMed] [Google Scholar]
- Fisher, B.M., Ha, J.H., and Raines, R.T. 1998. Coulombic forces in protein-RNA interactions: Binding and cleavage by ribonuclease A and variants at Lys7, Arg10, and Lys66. Biochemistry 37 12121–12132. [DOI] [PubMed] [Google Scholar]
- Guex, N. and Peitsch, M.C. 1997. SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis 18 2714–2723. [DOI] [PubMed] [Google Scholar]
- Hayashi, R., Moore, S., and Merrifield, R.B. 1973. Preparation of pancreatic ribonuclease 1–114 and 1–115 and their reactivation by mixture and synthetic COOH-terminal peptides. J. Biol. Chem. 248 3889–3892. [PubMed] [Google Scholar]
- Jones, T.A., Zou, J.Y., Cowan, S.W., and Kjeldgaard, M. 1991. Improved methods for the building of protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47 110–119. [DOI] [PubMed] [Google Scholar]
- Klink, T.A., Woycechowsky, K.J., Taylor, K.M., and Raines, R.T. 2000. Contribution of disulfide bonds to the conformational stability and catalytic activity of ribonuclease A. Eur. J. Biochem. 267 566–572. [DOI] [PubMed] [Google Scholar]
- Kraulis, P.J. 1991. MOLSCRIPT: A program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24 946–950. [Google Scholar]
- Laskowski, R.A., MacArthur, M.W., Moss, D.S., and Thornton, J.M. 1993. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26 283–291. [Google Scholar]
- Liang, J., Edelsbrunner, H., and Woodward, C. 1998. Anatomy of protein pockets and cavities: Measurement of binding site geometry and implications for ligand design. Protein Sci. 7 1884–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewenthal, R., Sancho, J., and Fersht, A.R. 1992. Histidine-aromatic interactions in barnase: Elevation of histidine pKa and contribution to protein stability. J. Mol. Biol. 224 759–770. [DOI] [PubMed] [Google Scholar]
- Martin, P.D., Doscher, M.S., and Edwards, B.F. 1987. The refined crystal structure of a fully active semisynthetic ribonuclease at 1.8-Å resolution. J. Biol. Chem. 262 15930–15938. [DOI] [PubMed] [Google Scholar]
- Miller, S., Janin, J., Lesk, A.M., and Chothia, C. 1987. Interior and surface of monomeric proteins. J. Mol. Biol. 196 641–656. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. and Minor, W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276 307–326. [DOI] [PubMed] [Google Scholar]
- Quirk, D.J., Park, C., Thompson, J.E., and Raines, R.T. 1998. His...Asp catalytic dyad of ribonuclease A: Conformational stability of the wild-type, D121N, D121A, and H119A enzymes. Biochemistry 37 17958–17964. [DOI] [PubMed] [Google Scholar]
- Raines, R.T. 1998. Ribonuclease A. Chem. Rev. 98 1045–1066. [DOI] [PubMed] [Google Scholar]
- Rasmussen, B.F., Stock, A.M., Ringe, D., and Petsko, G.A. 1992. Crystalline ribonuclease A loses function below the dynamical transition at 220 K. Nature 357 423–424. [DOI] [PubMed] [Google Scholar]
- Ratnaparkhi, G.S. and Varadarajan, R. 1999. X-Ray crystallographic studies of the denaturation of ribonuclease S. Proteins 36 282–294. [PubMed] [Google Scholar]
- Rico, M., Santoro, J., Bermejo, F.J., Herranz, J., Nieto, J.L., Gallego, E., and Jimenez, M.A. 1986. Thermodynamic parameters for the helix-coil thermal transition of ribonuclease-S-peptide and derivatives from 1H-NMR data. Biopolymers 25 1031–1053. [DOI] [PubMed] [Google Scholar]
- Schultz, L.W., Hargraves, S.R., Klink, T.A., and Raines, R.T. 1998a. Structure and stability of the P93G variant of ribonuclease A. Protein Sci. 7 1620–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz, L.W., Quirk, D.J., and Raines, R.T. 1998b. His...Asp catalytic dyad of ribonuclease A: Structure and function of the wild-type, D121N, and D121A enzymes. Biochemistry 37 8886–8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker, K.R., Fairman, R., Schultz, D.A., Robertson, A.D., York, E.J., Stewart, J.M., and Baldwin, R.L. 1990. Side-chain interactions in the C-peptide helix: Phe8-His12+. Biopolymers 29 1–11. [DOI] [PubMed] [Google Scholar]
- Takano, K., Yamagata, Y., Fujii, S., and Yutani, K. 1997. Contribution of the hydrophobic effect to the stability of human lysozyme: Calorimetric studies and X-ray structural analyses of the nine valine to alanine mutants. Biochemistry 36 688–698. [DOI] [PubMed] [Google Scholar]
- Tanimizu, N., Ueno, H., and Hayashi, R. 1998. Role of Phe120 in the activity and structure of bovine pancreatic ribonuclease A. J. Biochem. 124 410–416. [DOI] [PubMed] [Google Scholar]
- Wlodawer, A., Bott, R., and Sjolin, L. 1982. The refined crystal structure of ribonuclease A at 2.0 Å resolution. J. Biol. Chem. 257 1325–1332. [PubMed] [Google Scholar]
- Zegers, I., Maes, D., Dao-Thi, M.H., Poortmans, F., Palmer, R., and Wyns, L. 1994. The structures of RNase A complexed with 3`-CMP and d(CpA): Active site conformation and conserved water molecules. Protein Sci. 3 2322–2339. [DOI] [PMC free article] [PubMed] [Google Scholar]