Abstract
The contribution of solvent-exposed charged residues to protein stability was evaluated using ubiquitin as a model protein. We combined site-directed mutagenesis and specific chemical modifications to first replace all Arg residues with Lys, followed by carbomylation of Lys-amino groups. Under the conditions in which all carboxylic groups are protonated (at pH 2), the chemically modified protein is folded and very stable (ΔG = 18 kJ/mol). These results indicate that surface charge–charge interactions are not an essential fundamental force for protein folding and stability.
Keywords: Protein stability, chemical denaturation, chemical modification, energetics, electrostatic interactions, circular dichroism spectroscopy, balance of forces
An important step to the solution of the protein folding problem is the detailed understanding of the balance of forces that stabilize the native protein structure (Yang et al. 1992; Fersht 1993; Lazaridis et al. 1995; Makhatadze and Privalov 1995; Matthews 1995; Pace et al. 1996). It is generally believed that proteins are stabilized by hydrophobic effect, hydrogen bonding, and packing (van der Waals) interactions. The large favorable contribution of these forces is offset by a large unfavorable entropy change on protein folding. The resulting stability of the protein, as measured by the Gibbs energy (ΔG), is only on the order of few tens of kilojoules. One type of interaction, the electrostatic interactions between ionizable residues, has not received a wide recognition as an important factor for protein stability. It was believed that surface residues are equally well exposed to the solvent in both the native and unfolded states and thus, should not contribute to the protein stability. As to the salt bridges, their contribution to protein stability appears to be highly context dependent (Anderson et al. 1990; Dao-Pin et al. 1991a; Hendsch and Tidor 1994; Marqusee and Sauer 1994). Recently, several groups reported experimental data that show a significant stabilization of proteins with substitutions of the surface charges (Loladze et al. 1999; Pace et al. 2000; Perl et al. 2000; Spector et al. 2000; Sanchez-Ruiz and Makhatadze 2001). These results posed a question about the significance and magnitude of the contribution of the surface charges to the stability of globular proteins. The only direct experimental way to answer this question is to measure the stability of a protein without any charges. This is challenging because a protein without charges is expected to have very low solubility, which makes overexpression in bacteria difficult. We decided to approach this problem from the chemical standpoint, that is, express and purify protein-containing charges and then use specific chemical modifications to eliminate some of the charges. We used the molecule of ubiquitin as a model globular protein for these experiments (Makhatadze et al. 1998; Loladze et al. 1999,2001; Thomas and Makhatadze 2000). In this protein all ionizable groups are fully solvent exposed. We first substituted all Arg residues with Lys residues, followed by treatment of the protein with potassium cyanate that specifically converts all basic lysine residues into neutral homocitrulline. The pH of the solution was then lowered below the pKa of carboxylic acid, rendering under these conditions a protein without any functional charges. Analysis of the structure and stability of this modified protein shows that its stability is equal to the stability of the unmodified protein. On the basis of these results we conclude that charged residues are not essential for protein folding or stability. However, this does not mean that the charged residues are worthless in context of protein folding and stability. They play at least two major roles: they define solubility of the protein and they could and are used to finely modulate protein stability.
Results and Discussion
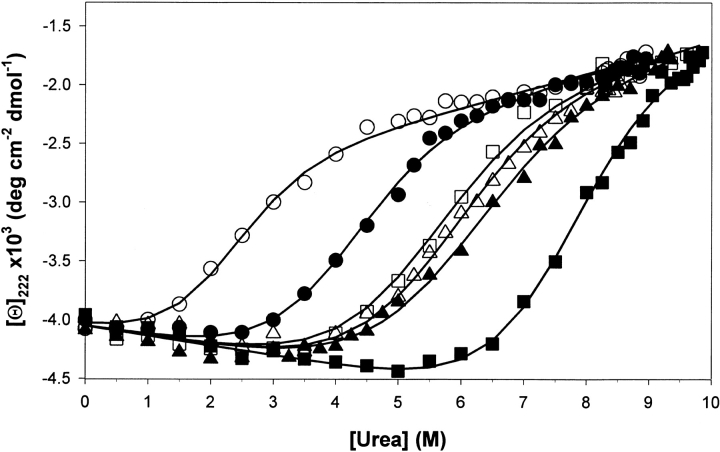
The ubiquitin molecule is maximally stable at neutral pH as estimated by temperature-induced unfolding using differential scanning calorimetry (Makhatadze et al. 1998). The stability at pH 6–8, however, is so high that it precludes experimental measurements of stability using temperature- or urea-induced unfolding (Loladze et al. 1999). The use of urea-induced unfolding is critical in our case because of the low solubility of chemically modified (CM) ubiquitin variants. The highest pH at which complete unfolding of all studied proteins by urea can be observed is pH 5.0. Figure 1 ▶ presents the results of urea-induced unfolding of ubiquitin variants monitored by the changes in the ellipticity at 222 nm. We used two ubiquitin variants, Ub11K and Ub10K, both of which have all Arg residues in the sequence substituted with Lys residues. Ub11K has the same number of ionizable residues as the wild-type protein. In the Ub10K, there is a single-site substitution at position 42, which is Glu as opposed to Lys in Ub11K. The unfolding profiles were analyzed using the linear extrapolation model according to equations 1 and 2 (see Materials and Methods). At pH 5.0 the Ub11K variant has midpoint of urea transition (C1/2) at 5.4 M and the Gibbs energy of unfolding in the absence of urea (ΔG°) is 16.8 kJ/mol. Under the same conditions the Ub10K variant has C1/2 = 7.6 M and ΔG° = 23.7 kJ/mol. This is consistent with our previous results showing that substitution for Glu is position 42 increases the stability by 6.8 kJ/mol (Loladze et al. 1999). This difference in stability can be qualitatively predicted using computational procedure that evaluates the energetic of charge–charge interactions (Loladze et al. 1999). The results of computation predict that the difference in the stabilities of ∼3 kJ/mol between Ub10K and Ub11K at pH 5 (Loladze et al. 1999) should decrease by only <10% upon pH decrease to 2. This is exactly what is observed experimentally. At pH 2 the Ub10K has unfolding transition characterized by C1/2 = 4.1 M and ΔG° = 12.8 kJ/mol, which is higher that that for Ub11K variant C1/2 = 2.2 M and ΔG° = 6.9 kJ/mol. Again, the large difference in stabilities (5.9 kJ/mol) between these two variants remains unchanged at this low pH. Of course, at pH 2, the carboxylic groups are largely protonated and the charge of the ubiquitin molecule is defined by the basic residues. Thus, upon carbamylation of lysine residues the difference in the stabilities between Ub10K and Ub11K should diminish. This is exactly what is observed (Fig. 1 ▶). At pH 2 the unfolding of CM Ub10K has ΔG° = 18.8 kJ/mol (C1/2 = 6.1 M) similar to that of CM Ub11K ΔG° = 17.7 kJ/mol (C1/2 = 5.7 M). The most striking is that the modified ubiquitin variants are extremely stable, even more stable than the unmodified Ub11K variant at pH 5.0. Thus, the ubiquitin molecule without functional charges is folded and extremely stable. According to the far-UV CD data, modified and unmodified ubiquitin variants are indistinguishable. The modifications did not affect the m value of urea-induced unfolding, thus indicating that at least according to this criterion (Dill and Shortle 1991) there are no changes in the unfolded state ensembles with chemical modifications. We can, therefore, conclude that surface charges are not required for folding and stability of the ubiquitin molecule. The question then is what is the role of charged residues for the structure and stability (we do not question the importance of charged residues for catalysis).
Fig. 1.
The dependence of ellipticity at 222 nm on the urea concentration for the unmodified and chemically modified ubiquitin variants at 25°C: Ub11K at pH 2.0 (○), Ub10K at pH 2.0 (•), Ub11K at pH 5.0 (□), Ub10K at pH 5.0 (▪), CM-Ub11K at pH 2.0 (▵), CM-Ub10K at pH 2.0 (▴). Solid lines show the results of the analysis using a linear extrapolation model (equation 1) with the parameters listed in the text.
Probably one of the most vital purpose for solvent-exposed charged residues is their importance for the solubility of proteins (Shaw et al. 2001). Intuitively, and from our direct experimental observations, the solubility of the ubiquitin variants changes by more than 50-fold (from >30 mg/mL to 0.7 mg/mL at pH 2) upon modification. However, the distribution of the charged residues cannot be random. Despite the fact that the charged residues are solvent exposed, charge–charge interactions can be both favorable and unfavorable (Loladze et al. 1999; Pace et al. 2000; Perl et al. 2000; Spector et al. 2000; Sanchez-Ruiz and Makhatadze 2001). Thus, the distribution of charged residues on the surface should be such that the overall energy of the charge–charge interactions is favorable or at the very least not destabilizing. In fact, this was noted previously by Spassov et al. (1994) and shown experimentally by others (Hollecker and Creighton 1982; Dao-Pin et al. 1991b). How does this reconcile with the recent experimental data suggesting optimization of charge–charge interactions as a way of increasing protein stability? There is no discrepancy between them. The protein structure appears to be stabilized by hydrophobic interactions, hydrogen bonding, and tight packing of the protein interior. Each of these factors contributes hundreds of kilojoules per mole to the Gibbs energy. These favorable stabilizing interactions are balanced by the large unfavorable entropy of protein folding. The resultant stability of the proteins is only several tens of kilojoules per mole. The energy of charge–charge interactions has the same order of magnitude. Because the charged residues are located mostly on the surface they do not affect significantly any of the other interactions and thus they can significantly affect intrinsic stability of the protein by amino acid substitution. This can be seen in Figure 1 ▶ for Ub10K and Ub11K, which differ in stability by 6.8 kJ/mol as a result of single amino acid substitution, and there are numerous other examples (see, e.g., Sanchez-Ruiz and Makhatadze 2001).
Materials and methods
Ubiquitin variants
The codons for Arg in the carboxy-terminally 6xHis-tagged yeast ubiquitin gene (Loladze et al. 2001) were replaced with the codons for Lys using a conventional PCR-based mutagenesis (Thomas and Makhatadze 2000). The incorporation of the mutations was confirmed by the sequencing of the entire gene using ABI 100 genetic analyzer. The mutations were incorporated in two backgrounds, one the wild-type yeast ubiquitin designated as Ub11K and the other in the background of the site variant R42E (Ub10K). Expression, purification, and characterization procedures for ubiquitin variants were identical to those used before (Loladze et al. 2001).
Chemical modification
All lysine residues in two ubiquitin variants Ub11K and Ub10K were converted to homocitrullin (Hct) using carbamylation reaction with cyanate as described (Rimon and Perlmann 1968). The 20-mL reaction mixture contained ubiquitin at concentration 1 mg/mL in 8 M of urea, 50 mM of sodium phosphate at pH 8.0. Potassium cyanate was added in small portions to a final concentration of 0.2 M during the span of 24 h at 30°C. The reaction was stopped by adding 1 mL of 1 M Tris at pH 8.0 followed by the dialysis against water. The initial characterization of the modified proteins was done using isoelctrofocusing on PhastSystem (Pharmacia, NJ). Single bands with mobility close to that of calibration standard with the pI = 4.55 (trypsin inhibitor) but much higher than calibration standard with the pI = 3.75 (methyl red dye) were observed indicating complete modification of both variants (calculated pI 4.5 and 4.4 for CM Ub11K and CM-Ub10K, respectively) have an apparent pI below 4.5. The final characterization of the carbomylated ubiquitin variants was done using Voyager (Applied Biosystems) MALDI-TOF biospectroscopy workstation. Single peaks corresponded to the molecular masses of 9755 Da for CM Ub11K variant and 9711 Da for CM Ub10K variant.
Circular dichroism spectroscopy and data analysis
Urea-induced unfolding of the ubiquitin variants was monitored by following the changes in ellipticity at 222 nm using JASCO J-715 spectropolarimeter (Loladze et al. 1999). Automated titration system based on Microlab 500 (Hamilton, Reno, NV) dispenser and 1-cm rectangular quartz cell similar to that described in Stites et al. (1995) was used. The protein sample (0.05 mg/mL in either 5 mM of glycine at pH 2.0 or 5 mM of sodium acetate at pH 5.0) was allowed to equilibrate for 20 min at each urea concentration. Analysis of the data was done according to the linear extrapolation model as described (Pace 1990; Santoro and Bolen 1992):
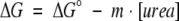 |
(1) |
where ΔG° is the Gibbs energy of unfolding in the absence of urea, m is the so-called m value that defines the dependence of the Gibbs energy, ΔG, on denaturant concentration (urea). The Gibbs energy at different concentrations of urea were calculated from the experimental data as:
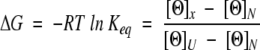 |
(2) |
where [Θ]N and [Θ]U are the ellipticities of the native and unfolded states, respectively, and the [Θ]x is the experimental ellipticity at a given concentration of urea. The data were fitted to equations 1 and 2 simultaneously as described in Loladze et al. (1999).
Acknowledgments
We thank Aimee van Olden for critical reading of the manuscript. This work was supported in part by grants from the NIH (GM54537) and NSF (MCB-0110396).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1101/ps.29902.
References
- Anderson, D.E., Becktel, W.J., and Dahlquist, F.W. 1990. PH-induced denaturation of proteins: A single salt bridge contributes 3–5 kcal/mol to the free energy of folding of T4 lysozyme. Biochemistry 29 2403–2408. [DOI] [PubMed] [Google Scholar]
- Dao-Pin, S., Sauer, U., Nicholson, H., and Matthews, B.W. 1991a. Contributions of engineered surface salt bridges to the stability of T4 lysozyme determined by directed mutagenesis. Biochemistry 30 7142–7153. [DOI] [PubMed] [Google Scholar]
- Dao-Pin, S., Soderlind, E., Baase, W.A., Wozniak, J.A., Sauer, U., and Matthews, B.W. 1991b. Cumulative site-directed charge–change replacements in basceriophage T4 lysozyme suggest that long-range electrostatic interactions contribute little to protien stability. J. Mol. Biol. 221 873–887. [DOI] [PubMed] [Google Scholar]
- Dill, K.A. and Shortle, D. 1991. Denatured states of proteins. Annu. Rev. Biochem. 60 795–825. [DOI] [PubMed] [Google Scholar]
- Fersht, A.R. 1993. The sixth Datta Lecture. Protein folding and stability: The pathway of folding of barnase. FEBS Lett. 325 5–16. [DOI] [PubMed] [Google Scholar]
- Hendsch, Z.S. and Tidor, B. 1994. Do salt bridges stabilize proteins? A continuum electrostatic analysis. Protein Sci. 3 211–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollecker, M. and Creighton, T.E. 1982. Effect on protein stability of reversing the charge on amino groups. Biochim. Biophys. Acta 701 395–404. [DOI] [PubMed] [Google Scholar]
- Lazaridis, T., Archontis, G., and Karplus, M. 1995. Enthalpic contribution to protein stability: Insights from atom-based calculations and statistical mechanics. Adv. Protein Chem. 47 231–306. [DOI] [PubMed] [Google Scholar]
- Loladze, V.V., Ibarra-Molero, B., Sanchez-Ruiz, J.M., and Makhatadze, G.I. 1999. Engineering a thermostable protein via optimization of charge–charge interactions on the protein surface. Biochemistry 38 16419–16423. [DOI] [PubMed] [Google Scholar]
- Loladze, V.V., Ermolenko, D.N., and Makhatadze, G.I. 2001. Heat capacity changes upon burial of polar and nonpolar groups in proteins. Protein Sci. 10 1343–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makhatadze, G.I. and Privalov, P.L. 1995. Energetics of protein structure. Adv. Protein Chem. 47 307–425. [DOI] [PubMed] [Google Scholar]
- Makhatadze, G.I., Lopez, M.M., Richardson, J.M. 3rd,, and Thomas, S.T. 1998. Anion binding to the ubiquitin molecule. Protein Sci. 7 689–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews, B.W. 1995. Studies on protein stability with T4 lysozyme. Adv. Protein Chem. 46 249–278. [DOI] [PubMed] [Google Scholar]
- Pace, C.N. 1990. Measuring and increasing protein stability. Trends Biotechnol. 8 93–98. [DOI] [PubMed] [Google Scholar]
- Pace, C.N., Shirley, B.A., McNutt, M., and Gajiwala, K. 1996. Forces contributing to the conformational stability of proteins. FASEB J. 10 75–83. [DOI] [PubMed] [Google Scholar]
- Pace, C.N., Alston, R.W., and Shaw, K.L. 2000. Charge–charge interactions influence the denatured state ensemble and contribute to protein stability. Protein Sci. 9 1395–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perl, D., Mueller, U., Heinemann, U., and Schmid, F.X. 2000. Two exposed amino acid residues confer thermostability on a cold shock protein. Nat. Struct. Biol. 7 380–383. [DOI] [PubMed] [Google Scholar]
- Rimon, S. and Perlmann, G.E. 1968. Carbamylation of pepsinogen and pepsin. J. Biol. Chem. 243 3566–3572. [PubMed] [Google Scholar]
- Sanchez-Ruiz, J.M. and Makhatadze, G.I. 2001. To charge or not to charge? Trends Biotechnol. 19 132–135. [DOI] [PubMed] [Google Scholar]
- Santoro, M.M. and Bolen, D.W. 1992. A test of the linear extrapolation of unfolding free energy changes over an extended denaturant concentration range. Biochemistry 31 4901–4907. [DOI] [PubMed] [Google Scholar]
- Spassov, V.Z., Karshikoff, A.D., and Ladenstein, R. 1994. Optimization of the electrostatic interactions in proteins of different functional and folding type. Protein Sci. 3 1556–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector, S., Wang, M., Carp, S.A., Robblee, J., Hendsch, Z.S., Fairman, R., Tidor, B., and Raleigh, D.P. 2000. Rational modification of protein stability by the mutation of charged surface residues. Biochemistry 39 872–879. [DOI] [PubMed] [Google Scholar]
- Stites, W.E., Byrne, M.P., Aviv, J., Kaplan, M., and Curtis, P.M. 1995. Instrumentation for automated determination of protein stability. Anal. Biochem. 227 112–122. [DOI] [PubMed] [Google Scholar]
- Thomas, S.T. and Makhatadze, G.I. 2000. Contribution of the 30/36 hydrophobic contact at the C-terminus of the alpha-helix to the stability of the ubiquitin molecule. Biochemistry 39 10275–10283. [DOI] [PubMed] [Google Scholar]
- Yang, A.S., Sharp, K.A. and Honig, B. 1992. Analysis of the heat capacity dependence of protein folding. J. Mol. Biol. 227 889–900. [DOI] [PubMed] [Google Scholar]