Abstract
Polyglutamine domains are excellent substrates for tissue transglutaminase resulting in the formation of cross-links with polypeptides containing lysyl residues. This finding suggests that tissue transglutaminase may play a role in the pathology of neurodegenerative diseases associated with polyglutamine expansion. The glycolytic enzyme GAPDH previously was shown to tightly bind several proteins involved in such diseases. The present study confirms that GAPDH is an in vitro lysyl donor substrate of tissue transglutaminase. A dansylated glutamine-containing peptide was used as probe for labeling the amino-donor sites. SDS gel electrophoresis of a time-course reaction mixture revealed the presence of both fluorescent GAPDH monomers and high molecular weight polymers. Western blot analysis performed using antitransglutaminase antibodies reveals that tissue transglutaminase takes part in the formation of heteropolymers. The reactive amino-donor sites were identified using mass spectrometry. Here, we report that of the 26 lysines present in GAPDH, K191, K268, and K331 were the only amino-donor residues modified by tissue transglutaminase.
Keywords: GAPDH, lysine residues, mass spectrometry, tissue transglutaminase
Transglutaminases (TGs, EC 2.3.2.13) are calcium-dependent enzymes that catalyze an acyl transfer reaction between the γ-carboxamide group of a peptide-bound glutamine residue and the ɛ-amino group of a peptide-bound lysine leading to an isopeptide bond (Folk and Finlayson 1977; Lorand and Conrad 1984). This reaction results in cross-linked protein polymers that are resistant to chemical and enzymatic attacks. Proteolytic enzymes able to hydrolyze these cross-links have not been identified in vertebrates (Melino and Piacentini 1998). At present, distinct widely expressed TG molecular forms have been described, including plasma factor XIIIa, keratinocyte TG (type I), tissue TG (type II, tTG), epidermal TG (type III), prostate TG (type IV; Aeschlimann and Paulsson 1994) and TG X (Kim et al. 1999). TG enzymes are involved in many fundamental biological processes including the fibrin clotting cascade (Folk and Finlayson 1977), seminal vesicle coagulation (Esposito et al. 1996), cornification of the epidermis, hair, and nails (Steinert et al. 1999), extracellular matrix and bone formation (Aeschlimann and Thomazy 2000), apoptosis (Melino and Piacentini 1998). Although the physiological function of these enzymes is to stabilize biological structures, there is increasing evidence that tTGs may play a role in the pathology of neurodegenerative diseases associated with (CAG)n expansion in the genome and to corresponding polyglutamine (Qn) repeats in the encoded protein that characterizes the disorder (Paulson 1999; Perutz 1999). There is a lot of evidence indicating that a common feature of the progressive neurodegenerative disorders is the accumulation of Qn-containing protein aggregates described in the nucleus and in the cytoplasm of affected neurons of patients, in mice models of CAG disease, and in cell-based systems (Davies et al. 1997; DiFiglia et al. 1997; Paulson et al. 1997; Gutekunst et al. 1999; Li et al. 1999). Several clinical and experimental findings suggest that aggregates are formed when the expansion of Q residues exceeds a critical number (n > 35–40), leading to a toxic gain of function that causes cell-specific neurodegeneration (Perutz 1999). The expanded Qn repeats may interact with each other through a polar zipper, thus contributing to aggregate formation (Perutz et al. 1994). On the other hand, it has been shown that proteins with Qn repeats are substrates of tTG, resulting in the formation of covalently bonded aggregates with polypeptides that contain lysyl residues (Green 1993; Kahlem et al. 1996; Cooper et al. 1997a,1999). These findings are supported by the observation that overexpression of the TG gene in human neuroblastoma cells greatly increase the number and size of cellular aggregates, whereas the inhibition of TG activity interferes with the accumulation of such cellular inclusions (de Cristofaro et al. 1999).
Kahlem and coworkers showed that the brain possesses K donor–containing proteins able to form covalent bridges with Qn domains (Kahlem et al. 1996). Subsequently, the glycolytic enzyme GAPDH was shown to tightly bind several proteins involved in Qn expansion disease: huntingtin, atrophin-1, ataxin-1, and the androgen receptor (Burke et al. 1996; Kosby et al. 1996) in human brain homogenates. In addition, GAPDH was found covalently linked to the Q60 sequence in a Balb-c 3T3 cell line overexpressing human tTG when the cell content was treated with the Q60 polymer (Gentile et al. 1998). These findings suggested that GAPDH possesses at least one reactive K residue per monomer forming bridges between two or more Qn stretches (Cooper et al. 1999). To date, no structural data are available to solve the issue of how many and which lysine residues in GAPDH are substrates for TG enzyme.
The work described in the present article shows that rabbit muscle GAPDH is a lysyl donor substrate of the guinea pig liver tTG. GAPDH was incubated in the presence of tTG and a Q-donor peptide, Substance P (SubP), which previously was shown to be a substrate of tTG (Esposito et al. 1995). The products of the reaction of GAPDH and SubP in the presence of tTG were characterized by SDS-PAGE and Western blot analyses. The reactive amino-donor sites were identified by mass spectrometry. This article reports that only three lysines, of the 26 lysine residues present in GAPDH, are involved in the formation of cross-links with SubP. These results suggest a high specificity of the tTG-catalyzed reaction.
Results
Glyceraldehyde 3-phosphate dehydrogenase is a substrate of tissue transglutaminase
GAPDH was incubated with dansylated amino-acceptor Substance P (RPKPQQFFGLM) (DNS-SubP) in the presence of purified tTG. The reaction was conducted for different time lengths after a time-course experiment (1, 4, and 18 h). The formation of insoluble products, clearly visible in the assay tubes, forced us to set up a reaction mixture for each incubation period, within a single experiment. The products of the tTG-catalyzed reaction were analyzed by 12% SDS-PAGE, fluorescence, and Western blot analyses.
The Coomassie-stained SDS-PAGE pattern shown in Figure 1A ▶ revealed the presence of the GAPDH monomer migrating at 36 kD in the reaction mixtures after 1, 4, and 18 h of incubation (Fig. 1A ▶, lanes 4, 5, and 6, respectively). At the same time, very high molecular weight species appeared from the first stages of reaction. Such polymers hardly penetrated the gel mesh for short reaction times (Fig. 1A ▶, lane 4) whereas for longer periods of incubation they could not leave loading gel wells because of their very large size (Fig. 1A ▶, lanes 5,6). The formation of polymers was inhibited by the addition of a calcium chelator (data not shown). Polymers were also present when SubP was omitted from the reaction (Fig. 1A ▶, lane 3). These results confirm that GAPDH contains reactive glutamines (Ikura et al. 1998) and lysines (Cooper et al. 1997a; Gentile et al. 1998) because intermolecular cross-links are formed in the presence of tTG.
Fig. 1.
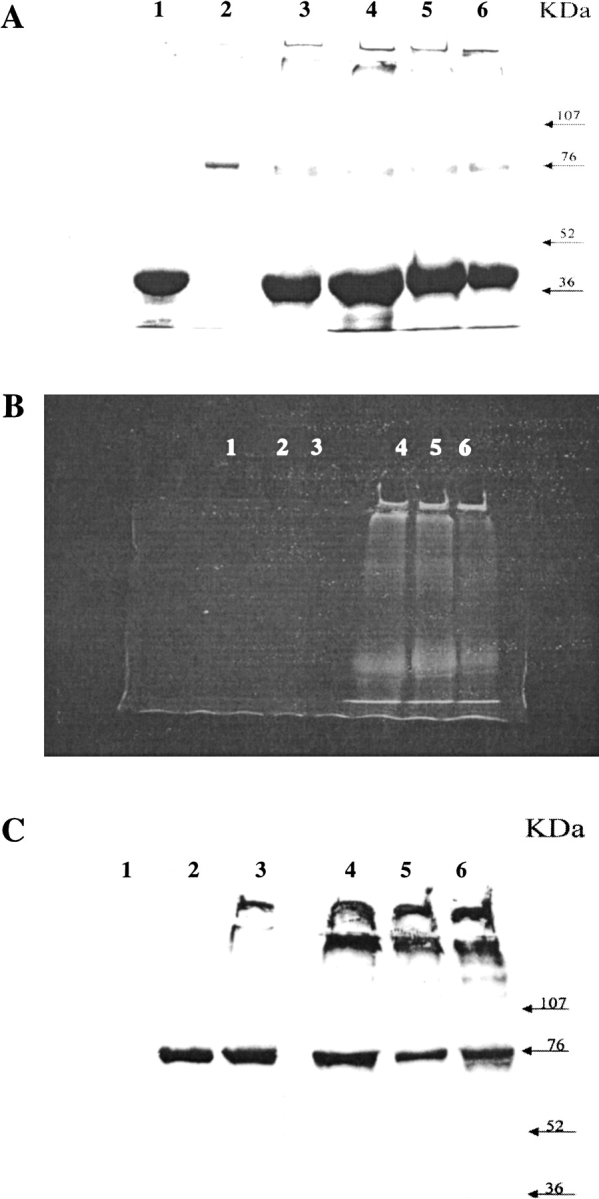
Substrate capacity of GAPDH in tTG-mediated cross-links. SDS-PAGE gel Coomassie blue stained (A), fluorescence (B) and corresponding Western Blot analysis performed using anti-tTG antibodies (C) of the in vitro incubation of rabbit muscle GAPDH (50 μg) with dansylated SubP (18 μg) in the presence of purified tTG (5 μg) at 37°C for 1, 4, and 18 h (lanes 4, 5, and 6, respectively). GAPDH and purified tTG were used as controls (lanes 1 and 2, respectively). As additional control, GAPDH was incubated at 37°C for 18 h in the presence of tTG (lane 3). Protein marker levels are indicated.
The presence of a fluorescent probe (DNS) in the DNS-SubP allowed us to monitor the occurrence of cross-links involving the dansylated peptide by fluorescence analysis. The fluorescence patterns of lanes 4–6, shown in Fig. 1B ▶, showed a very intense band at 36 kD corresponding to DNS-SubP linked to the GAPDH monomer. The latter showed the same migration behavior in the SDS-PAGE (see Fig. 1A ▶). An intense fluorescent band also was observed at very high molecular weight because of species characterized by very large size that hardly penetrated the gel mesh in the early stages of the reaction (Fig. 1B ▶, lane 4). These species could not leave the loading gel wells in the late stages (Fig. 1B ▶, lanes 5, 6), as previously observed in the SDS-PAGE analysis. These results show the involvement of DNS-SubP in the formation of high molecular weight heteropolymers containing GAPDH as a tTG amino donor.
Western blot analysis performed using anti tTG antibodies (Fig. 1C ▶) showed an intense band at a high molecular weight. This finding reveals that tTG takes part in the formation of high molecular weight heteropolymers together with DNS-SubP and GAPDH.
Identification of reactive lysine residues by mass spectrometric analysis
The SDS-PAGE bands at 36 kD (Fig. 1A ▶, lanes 4–6) were excised and separately submitted to an in-gel hydrolysis to identify the K residues involved in the covalent cross-links with DNS-SubP. An involvement of GAPDH Q residues in the formation of cross-links was not expected because the K residue in SubP was reported previously not to be a tTG substrate (Porta et al. 1988).
The proteolytic step was conducted on each band, using endoproteinase Asp-N sequencing grade, and the resulting peptide mixtures were analyzed by MALDI-MS. The proteolytic enzyme was chosen because of its ability to hydrolyze GAPDH and to produce a suitable peptide mixture, leaving the DNS-SubP intact. The reasoning behind this choice was to facilitate the assignment of peptides linked to intact DNS-SubP. As a control, the amino acid sequence of GAPDH was characterized using the same strategy as described in Materials and Methods.
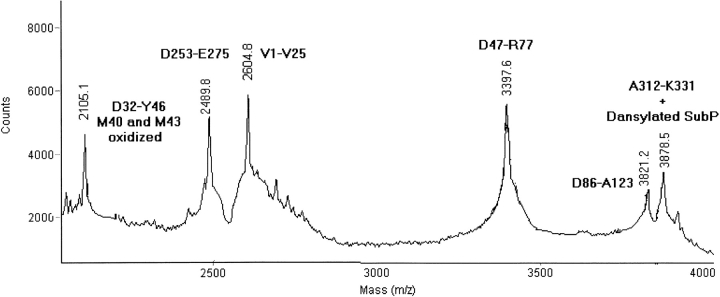
MALDI-MS spectra of the peptide mixtures, produced by the hydrolysis of the incubation product at 1, 4, and 18 h, showed signals that could be assigned to GAPDH fragments on the basis of their molecular weights and the specificity of the proteolytic enzyme. Only a few mass signals could not be related to peptides along the GAPDH sequence. However, they were interpreted as GAPDH fragments covalently linked to DNS-SubP via an isopeptide bond. Figure 2 ▶ shows a partial MALDI-MS spectrum of the peptide mixture obtained from the hydrolysis of the assay after 1 h. Signals at m/z 2105.1, 2489.8, 2604.8, 3397.6, and 3821.2 were assigned to peptides along the GAPDH sequence on the basis of their molecular weights. An additional signal at m/z 3878.5 was detected and assigned to peptide A312-K331 covalently linked to DNS-SubP (expected m/z 3878.2). The sequence of the identified GAPDH fragment meant K331 could be identified unequivocally as a tTG amino-donor site. Table 1 shows the mass signals due to the formation of cross-links between GAPDH peptides and DNS-SubP at different incubation times. The signals at m/z 3879.0 and 3879.9 were also observed at 4 and 18 h, thus confirming the occurrence of a cross-link involving K331 as an amino donor. An additional signal at m/z 2579.8 was detected in the 4-h mixture and assigned to peptide D186-R194 linked to DNS-SubP (expected m/z 2579.7). Peptide D186-R194 contains a single K residue at position 191, which therefore is involved in the cross-link formation. The presence of this signal only after 4 h seems to suggest that K191 is less reactive than K331. The absence of the signal 2579.7 after a longer incubation time (18 h) can be caused by low recovery of the peptides extracted from the gel. The characterization of reactive lysines in solution (see below) showed, in fact, the presence of this signal also at 18 h.
Fig. 2.
Partial MALDI-MS spectrum of the peptide mixture derived by Asp-N hydrolysis of the 1-h incubation mixture. The assignment of peaks to the corresponding peptides is shown.
Table 1.
MALDI-MS data of the AspN peptide mixtures generated by in gel hydrolysis of the 36kD bands corresponding to in vitro assay at different times of incubation
| 1h | 4h | 18h | Assignment | Reactive K |
| 2579.8 | (D186-R194)+DNS-SubP | 191 | ||
| 3878.5 | 3879.0 | 3879.9 | (D312-K331)+DNS-SubP | 331 |
The reported characterization highlighted the role of GAPDH as an amino donor and led to the location of the K residues involved in the tTG catalyzed cross-links. To confirm and extend these results, we repeated the incubation by using unmodified SubP for different time lengths corresponding to 5 min, 30 min, 1 h, 4 h, and 18 h. Every mixture formed during the reaction containing soluble and nonsoluble products was hydrolyzed by endoproteinase AspN, and the resulting peptide mixtures were analyzed by MALDI-MS. Table 2 shows the mass signals caused by the formation of cross-links between GAPDH peptides and SubP at different incubation times. A signal at m/z 2347.0 was assigned to peptide D186-R194 covalently linked to a SubP molecule (expected m/z, 2347.7), thus confirming a previous finding that K191 is a tTG amino-donor site. Signals at m/z 3774.4 and 2492.3 were assigned, respectively, to GAPDH peptides D312-E332 and D323-E332 linked to a SubP molecule (expected m/z 3774.4 and 2492.0, respectively), thus confirming that K331 is indeed a tTG amino-donor site.
Table 2.
MALDI-MS data of the peptide mixtures generated by Asp-N hydrolysis of the in vitro assays after 5 min, 30 min, 1 h, 4 h, and 18 h of incubation
| MH+ | Assignment | Reactive K | 5` | 30` | 1h | 4h | 18h |
| 2347.0 | (D186-R194)+SubP | K191 | n.d. | n.d. | n.d. | √ | √ |
| 2492.3 | (D323-E332)+SubP | K331 | √ | √ | √ | √ | √ |
| 2609.4 | (E264-E275)+SubP | K268 | √ | √ | √ | √ | √ |
| 3820.6 | (D253-E275)+SubP | K268 | √ | √ | √ | √ | √ |
| 3774.4 | (D312-E332)+SubP | K331 | √ | √ | √ | √ | √ |
(√) Signal detected.
(n.d.) Signal not detected.
Reactive K residues are shown.
The structural characterization in solution led to the identification of one other K residue sensitive to tTG catalysis. The signal at m/z 2609.4 was, in fact, attributed to the GAPDH peptide E264-E275 bound to a SubP molecule via an isopeptide bond that involved K268 (expected m/z, 2609.0). MALDI spectra also showed a signal at m/z 3820.6, which was assigned to the peptide D253-E275 covalently linked to SubP (expected m/z 3821.4). The latter GAPDH fragment contained four putative tTG substrates (K256, K257, K260, and K268). The presence of a mass signal corresponding to the unmodified peptide D253-E263, and the absence of the modified peptide D253-E263, led to the identification of K268 as a tTG-sensitive residue, thus confirming the assignment of the mass signal at m/z 2609.4. The comparison of MALDI-MS spectra corresponding to different incubation periods, shows that the three lysine residues identified as tTG substrates were not kinetically equivalent in the reaction. K268 and K331 were involved in the formation of the cross-links from the very beginning of incubation, whereas K191 was detected only after 4 h of incubation. Note that the absence of signals corresponding to the formation of cross-links involving K268 in the previous in-gel analysis was probably related to the already mentioned low recovery of peptides extracted from the gel.
Finally, it is important to consider that the peptide mixtures contained both modified and unmodified forms of the GAPDH fragments, indicating that the selective enzymatic modification of lysine residues is not complete, that is, some GAPDH molecules contain unmodified K residues whereas others contain the same residues modified by tTG catalyzed reaction.
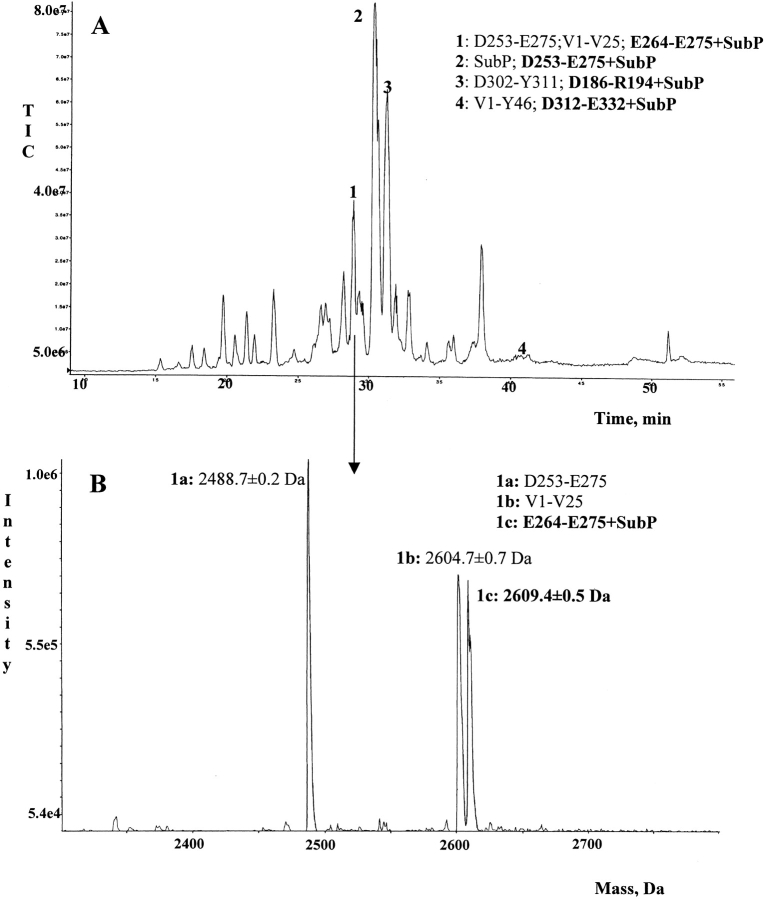
To confirm the previous data, the 4-h mixture was separated by HPLC using a C18 reverse phase column and analyzed on-line by electrospray mass spectrometry (ESI-MS). The total ion current (TIC) profile of the peptide mixture is reported in Figure 3A ▶. Every TIC fraction is correlated to a mass spectrum; the analysis of each mass spectrum provided unambiguous confirmation that K191, K268, and K331 are tTG substrates as shown by previous experiments. The TIC fractions containing GAPDH peptides modified by SubP are numbered in Figure 3A ▶. Figure 3B ▶ shows the mass spectrum correlated to fraction 1 as an example. The mass spectrum shows the presence of three different peptides, one of them being the fragment E264-E275 covalently linked to SubP molecule.
Fig. 3.
(A) LC/ESI-MS analysis of the peptide mixture obtained from endoproteinase Asp-N hydrolysis of the 4-h incubation mixture. Fractions containing modified GAPDH peptides are numbered. Peptides present in the numbered fractions are shown. Peptides containing cross-links with SubP are shown in boldface. (B) Transformed ES mass spectrum of fraction 1. Assignments of the mass signals to the corresponding peptides are shown.
Effect of NAD+
In vivo GAPDH acts as a homotetramer that binds NAD+ (Sirover 1999). In vitro assays were performed in the presence of NAD+ to verify whether it could affect the reactivity of K residues as tTG substrates. The samples were treated as described for the experiments in the absence of NAD+. The analyses of AspN peptide mixtures by MALDI-MS showed that NAD+ did not affect the number or the location of K residues sensitive to tTG activity. K191, K268, and K331 were, in fact, the only amino-donor amino acids modified by the enzymatic reaction even in the presence of the cofactor.
Localization of lysine residues in the three-dimensional structure of GAPDH
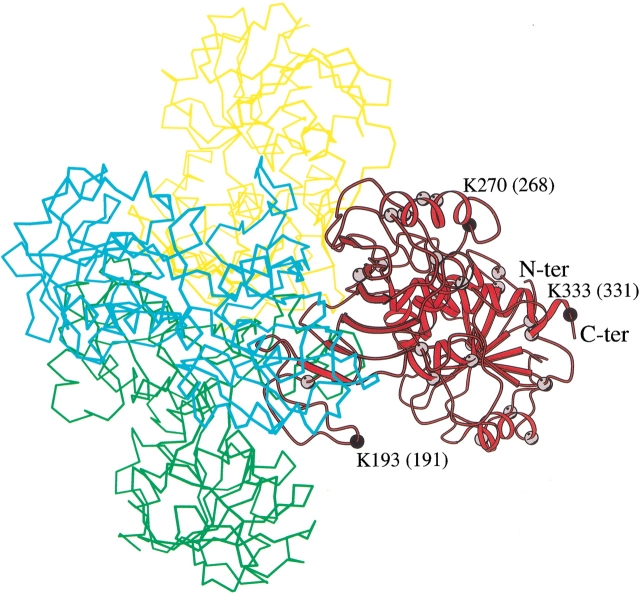
Over the years, several crystallographic structures of GAPDH from different sources have been reported. All these models share a high structural similarity. In fact, GAPDH is a tetrameric enzyme the subunits of which have a cofactor binding site and a catalytic domain. To identify the position of the lysine residues in the three-dimensional structure of rabbit GAPDH, we used the structure of human skeletal muscle GAPDH (Mercer et al. 1976) as a model, because the sequences of rabbit and human enzymes are very similar (90% of the residues are identical). Molecular graphics analysis shows that, with the sole exception of K306 located at the intersubunit interface, all the lysines are solvent exposed (Fig. 4 ▶). This observation is in agreement with the results obtained by Lambert and Perham (1977) who showed that only one amino group per subunit of rabbit GAPDH was completely unavailable for the reaction with methyl acetamidate.
Fig. 4.
GAPDH tetramer drawn using the coordinates of the Protein Data Bank entry code 3GDP (Mercer et al. 1976). A ribbon representation has been used to draw one subunit. The balls represent the location of lysine residues in this subunit. The number of the reactive lysine residues (in black) refers to the human sequence. The number of reactive lysine residues along the rabbit sequence is given in parenthesis. The trace of the other subunits is also shown. The figure was drawn using MOLSCRIPT (Kraulis 1991).
Furthermore, the molecular graphics analysis reveals that the reactive K191, K268, and K331 belong to rather flexible regions of the protein. In fact, K191 and K268 are located in loop regions whereas K331 is the penultimate residue of the C-terminal end of the enzyme.
Discussion
The accumulation of glutamine-repeat containing protein aggregates within the cytosol and nuclei of affected neurons appears to be a crucial step in the pathogenesis of progressive neurodegenerative disorders, such as Huntington disease (HD;Davies et al. 1997; DiFiglia et al. 1997; Paulson et al. 1997; Gutekunst et al. 1999; Li et al. 1999). Nuclei from dying neurons in the basal ganglia and the brain cortex of transgenic animals with the mutated huntingtin gene have been shown to contain large, insoluble high molecular protein aggregates (Davies et al. 1997). It has been reported that polyglutamine-expanded proteins are able to associate both to themselves and to specific proteins. For example, huntingtin associates with polymerized microtubules in vitro (Tukamoto et al. 1997), and ataxin-1 binds to leucine-rich acidic nuclear protein (Matilla et al. 1997). It has been hypothesized that a polar zipper model serves as a nucleation site for noncovalent interactions in fibrillous aggregate formation of huntingtin and other proteins (Perutz et al. 1994; Liu et al. 2001). The same author, who initially put forward the hypothesis that huntingtin aggregation could take place through the polar zipper action (Perutz et al. 1994), more recently (Perutz 1999) suggested that aggregate formations also could occur through cross-links catalyzed by tTG or by a combination of both mechanisms. In fact, it was shown that polyglutamine domains are good substrates of the enzyme tTG in vitro, and that larger pathological-length Qn domains are the most active substrates of the enzyme (Cooper et al. 1997a).
At present, there is little information concerning the amino-donor partner of polyglutamine containing proteins to form covalently bonded aggregates (Kahlem et al. 1996; Cooper et al. 2000). It has been shown that the glycolytic enzyme GAPDH binds tightly to several disease proteins such as huntingtin, atrophin-1, ataxin-1, and the androgen receptor (Burke et al. 1996; Kosby et al. 1996). The finding that four different proteins, implicated in different neurodegenerative diseases, interact with GAPDH suggests that common biochemical mechanisms involving GAPDH may lead to different pathogenesis.
On the basis this, we have undertaken a study aimed at structurally characterizing the products of the tTG catalyzed reaction involving GAPDH. We showed that GAPDH is a lysyl donor substrate of tTG in vitro. Surprisingly, of 26 lysines present in the GAPDH sequence (Lambert and Perham 1977), only K191, K268, and K331 are sites of tTG-dependent cross-link formation. In principle, the observed specificity could be for several possible reasons: (1) the sequence around the NH2 donor, (2) the solvent accessibility of lysine side chains, and (3) steric hindrance between tTG and GAPDH, which may prevent the tTG recognition of specific lysine residues.
It is well known that it has not been possible to derive a consensus sequence around the specific lysine residues from the few substrate proteins in which the amino-donor sites have been identified (Pucci et al. 1988; Porta et al. 1991; Mariniello et al. 1993; Grootjans et al. 1995). Nevertheless, it has been shown that the nature of the amino acid residues directly preceding the lysine may influence its reactivity. Indeed, uncharged, basic polar and small aliphatic residues enhance reactivity, which is decreased by residues such as D, G, P, H, and W (Grootjans et al. 1995). In this context, the observed reactivity of GAPDH K331, which is preceded by a serine residue, is in line with this trend (Groenen et al. 1994; Grootjans et al. 1995). The reactivity of K331 also is enhanced by its location in the C-terminal end of the protein. These results support the general pattern seen so far for other substrate proteins that essentially head and tail domain sequences participate in cross-link reactions (Steinert et al. 1999).
The reactivity of K268, located in the L-K-G sequence, is not surprising because it has been shown that a leucine residue directly adjacent to the lysine substrate has a positive effect on its reactivity (Groenen et al. 1994). The reactivity of GAPDH K191, which is preceded by a glycine, is certainly surprising, as it has been shown that this residue has an adverse effect on the tTG reaction (Grootjans et al. 1995). To the best of our knowledge, this is the first observed case in which a K residue, preceded by a glycine residue, turns out to be a tTG substrate. However, note that other GAPDH lysine residues are not amino donors, despite the fact that they are located in regions with sequences that should have enhanced their reactivity. Altogether, these observations show that previous data on the effect of the local structure on lysine reactivity can only partially explain the specificity of the cross-linked site of GAPDH.
The analysis of the three-dimensional model of GAPDH suggests that 25 of 26 lysines are solvent exposed, and therefore they can be potential amino donors. In fact, it has been reported that these lysines are reactive toward chemical reagents (Lambert and Perham 1977). In this scenario, the selectivity of the GAPDH lysine residues in the reaction catalyzed by tTG could be mostly caused by steric hindrance, which rules the recognition of lysine residues by tTG.
The catalytic site of GAPDH is localized in the C-terminal region (Sirover 1999). Our findings that all the reactive lysines are present in the C-terminal portion of the molecule may explain the previous observation that purified GAPDH was inactivated by tTG in the presence of glutathione S-transferase constructs containing a Qn domain of pathological length (Cooper et al. 1997b). This observation suggested that loss of GAPDH activity might contribute to the decrease of energy metabolism in HD brain (Burke et al. 1996). However, GAPDH is expressed in enormous amounts relative to huntingtin, and it is improbable that binding of huntingtin to GAPDH could make much difference to the level of the glycolytic enzyme. Conversely, GAPDH, which was studied for more than 3 decades for its pivotal role in glycolysis, has multifunctional activities and diverse subcellular localizations. These include roles for GAPDH in membrane transport and fusion, microtubule assembly, DNA replication, and DNA repair (Sirover 1999). In addition, GAPDH may play a role in apoptosis, which specifically involves its translocation to the nucleus (Saunder et al. 1997; Sawa et al. 1997). Sequence analysis suggests the possibility that amino acids within the catalytic domain, containing the tTG reactive lysine residues, may be involved in the regulation of the subcellular localization of GAPDH in mammalian cells (Yoneda 1997). Therefore, it is conceivable that TG-catalyzed post-translational modification of GAPDH may influence its nuclear translocation as well as any of its other several functions.
In conclusion, we set up an easy and reliable strategy for the identification of tTG-reactive lysine residues in GAPDH. Although we used SubP that has no involvement in HD pathology, the strategy can be effectively used to study the aggregation processes with extended polyQ domains containing normal and pathological Q repeats.
Materials and methods
Materials
Rabbit muscle GAPDH, guinea pig liver tissue transglutaminase (tTG), Substance P (SubP), NAD+, dithiothreitol (DTT), α-cyano-4-hydroxycinnamic acid, and SDS-PAGE protein standards were purchased from Sigma. Iodoacetamide (IANH2) was obtained from Fluka and endoproteinase Asp-N sequencing grade from Roche Diagnostics GmbH. RP-HPLC columns C18 (25 × 0.46 cm) and (25 × 0.21 cm) were purchased from Vydac (The Separations Group). All reagents for electrophoresis application were purchased from Bio-Rad. Pipette Zip-Tip C18 for sample preparation were obtained from Millipore. 1-Dimethylamino naftalen solphonil chloride (dansylchloride, DNS-Cl) and all other reagents and solvents were of the highest purity available from Carlo Erba.
tTG was purified as previously described (Lee et al. 1989).
Structural characterization of GAPDH
An aliquot of GAPDH corresponding to 35 μg (1 nmole) was denatured in 6 M Guanidine-HCl, 0.25 M Tris-HCl, 1.25 mM EDTA at pH 8.5 and reduced with DTT using a reagent molar excess of 10 : 1 versus cysteine moles. The reaction was performed for 2 h at 37°C. The reduced and denatured GAPDH then was alkylated by incubation with iodoacetamide. The alkylating agent was added to the protein solution using a −SH/IANH2 = 1 : 10 molar ratio. The reaction was conducted at room temperature for 1 h and then halted by RP-HPLC desalting. Chromatographic analysis was performed using a Vydac C4 column (25 × 0.46 cm) and an elution system consisting of 0.1% TFA (eluent A) and 0.07% TFA in 95% acetonitrile (eluent B). The modified protein was eluted by means of a linear gradient of eluent B from 10% to 80% over 40 min, at a flow rate of 1 mL/min. Elution was monitored at 220 nm, and the collected fractions were analyzed by ESI-MS. The fraction containing the alkylated GAPDH was lyophilized and then dissolved in 50 mM ammonium bicarbonate at pH 8.1. The protein then was submitted to enzymatic hydrolysis using endoproteinase Asp-N. The reaction was performed for 18 h at 37°C and stopped by adding 5 μL of 5% TFA. The peptide mixture was analyzed by MALDI-MS, and mass signals were assigned to GAPDH fragments on the basis of their molecular weights and the specificity of the enzyme.
The structural analysis meant that the whole sequence of GAPDH from rabbit muscle could be verified. Note that it was possible to identify the amino acid at position 123 as alanine, reported as an X in the SWISS-PROT sequence (accession number P46406). This experimental evidence is supported by the finding of Ala 123 in the GAPDH sequence from several other sources such as pig muscle (Harris and Perham 1968), lobster muscle (Davidson et al. 1967), ox liver (Heinz and Kulbe 1970), and Bacillus stearothermophilus (Walker et al. 1980).
Synthesis of DNS-SubP
Substance P (100 μg) was dissolved in 100 μL of 0.2 M NaHCO3 at pH 8.8 and incubated with 200 μL of a 5-mg/mL DNS-Cl acetone solution. The reaction was performed in the dark at 37°C. After 3 h, when the starting yellow mixture turned colorless, the reaction was stopped and the DNS-SubP was purified by RP-HPLC analysis using a C18 column (25 × 0.46 cm). The DNS-SubP was eluted using the same eluents described above, by means of a linear gradient of solvent B from 5% to 95% over 60 min, at a flow rate of 1 mL/min. Elution was monitored at 220 nm, and individual collected fractions were identified by ESI-MS.
tTG-catalyzed modification of lysine residues
Modification of K residues in GAPDH was performed by incubating 50 μg of the protein with 18 μg of SubP (molar ratio GAPDH/SubP = 1 : 10) in the presence of 5 μg of tTG (molar ratio GAPDH/tTG = 20 /: 1) in 125 mM Tris-HCl at pH 8.1, 10 mM DTT, 2.5 mM CaCl2 (final volume 32 μL). The experiments performed in the presence of NAD+ were set up by adding a GAPDH/NAD+ = 1 : 1 molar ratio. Each assay mixture was incubated for 5 min, 30 min, 1 h, 4 h, or 18 h at 37°C. Eighteen-hour incubations were provided with an additional 5-μg aliquot of tTG after 4 h. The reactions were stopped by lowering the pH (5 μL of TFA 5%) followed by lyophilization. Similar experiments were performed using DNS-SubP and then characterized by means of electrophoresis analyses (SDS-PAGE, fluorescence assay and Western blot analysis).
SDS-PAGE
The lyophilized samples were resuspended in 15 μL of sample buffer and analyzed by SDS-PAGE. SDS gel electrophoresis was performed on 12% linear slab gels as described by Laemmli (Laemmli 1970). Protein bands were stained by means of a Coomassie Blue Brilliant treatment. Molecular masses of protein bands were estimated by comparing their electrophoretical mobility to that of the molecular weight markers.
Fluorescence assay
The assay was performed on the samples analyzed by SDS-PAGE. Samples were incubated in a solution containing 50 mM Tris-HCl at pH 7.1, 9 M urea, 2% SDS, 40 mM DTT for 1 h before loading the mixtures on the stacking gel wells. After the electrophoresis run, the gel was rinsed in the dark in a 10% acetic acid, 25% 2-propanol solution for 15 min. Afterward, a picture of the gel was taken in the dark using a transilluminator (Tulp 20 M; Genenco).
Western blot
Protein bands were electroblotted onto a nitrocellulose strip. Binding of primary antibody, directed against tTG, was revealed using horseradish peroxidase-conjugate goat anti-rabbit IgG and 4-chloro-1-naphtol/hydroxyperoxide as chromogenic agents.
Enzymatic hydrolysis
Lyophilized samples were resuspended in 50 mM ammonium bicarbonate at pH 8.0. Enzymatic digestions with endoproteinase Asp-N were performed using a 1 : 100 enzyme-to-substrate ratio (w/w). Acetonitrile (10%) was added to activate the enzyme. Reactions were conducted for 18 h at 37°C and stopped by adding 5% TFA.
In-gel digestions of proteins were performed on the Coomassie blue–stained protein-containing bands excised from the gel. Gel pieces were washed with acetonitrile and then with 0.1 M ammonium bicarbonate. Protein samples were reduced by incubation in 10 mM DTT for 45 min at 56°C and alkylated with 55 mM iodoacetamide in 0.1 M ammonium bicarbonate for 30 min at room temperature, in the dark, as previously described (Esposito et al. 2001). The gel particles were then washed with ammonium bicarbonate and acetonitrile. Enzymatic digestions were conducted with endoproteinase Asp-N sequencing grade 15 ng/μL in 50 mM ammonium bicarbonate at pH 8.5 at 4°C for 4 h. The enzyme/buffer solution then was removed and a new aliquot of the buffer solution was added for 18 h at 37°C. A minimum reaction volume sufficient for complete rehydration of the gel was used. Peptides then were extracted washing the gel particles with 20 mM ammonium bicarbonate and 0.1% TFA in 50% acetonitrile at room temperature and then lyophilized.
Each peptide mixture was analyzed by MALDI-MS. In some cases, the samples were previously desalted using C18-Zip-Tip from Millipore.
Mass spectrometry
MALDI mass spectra were recorded using a PerSeptive Biosystem Voyager DE Instrument. A mixture of 1 μL of sample solution and 1 μL of α-cyano-4-hydroxycinnamic acid (10 mg/mL in acetonitrile/0.2% TFA = 7/3, v/v) was applied to the sample plate and allowed to dry. Mass calibration was performed using insulin (average molecular mass 5734.6 Daltons) and a matrix peak (379.1 Daltons) as internal standards. Raw data were analyzed using computer software provided by the manufactures and reported as average masses.
ESI-MS analyses were conducted using a Bio-Q triple quadrupole mass spectrometer (Micromass, Manchester, UK) equipped with an electrospray ion source. Samples were injected into the ion source (kept at 80°C) at a flow rate of 5μL/min using a syringe pump Harvard Apparatus mod. 11. Data were acquired and elaborated using the Masslynx program, purchased from Micromass. Mass calibration was performed by means of multiply charged ions from a separate injection of horse heart myoglobin (Sigma; average molecular mass: 16951.5 Daltons); all masses are reported as average values.
LC/MS analysis was performed using an API100 mass spectrometer (PerSeptive Biosystem) equipped with an electrospray ion source. Thirty microliters peptide mixture was injected onto a RP-HPLC C18 column (25 × 0.21 cm) and fractionated by performing a linear gradient from 5% to 65%B in 60 min, at a flow rate of 200 μL/min. The eluate then was split 1/5 before entering into the ion source. Spectra were acquired from 450 to 2000 Daltons. The resulting mass data were elaborated using the BioMultiview software provided by the manufacturer. The mass range was calibrated using horse heart myoglobin.
Acknowledgments
Dedicated to the memory of our friend and colleague Guido Sodano, Professor of Organic Chemistry. This work was supported by CNR grant CNRG0072CF, Progetto Giovani-Agenzia 2000.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Abbreviations
Asp-N, endoproteinase Asp-N
DNS, dansyl group, ESI-MS, electrospray mass spectrometry
HD, Huntington disease
LC/MS, liquid chromatography/mass spectrometry
MALDI-MS, matrix-assisted laser desorption ionization mass spectrometry
Qn, polyglutamine repeat containing n Q residues
RP-HPLC, reverse phase high pressure liquid chromatography
SubP, substance P
TG, transglutaminase
TIC, total ion current
tTG, guinea pig liver tissue transglutaminase
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1101/ps.17102.
References
- Aeschlimann, D. and Paulsson, M. 1994. Transglutaminases: Protein crosslinking enzymes in body fluids. Thromb. Haemost. 71 402–414. [PubMed] [Google Scholar]
- Aeschlimann, D. and Thomazy, V. 2000. Protein crosslinking in assembly and remodelling of extracellular matrices: The role of transglutaminases. Connect Tissue Res. 41 1–27. [DOI] [PubMed] [Google Scholar]
- Burke, J.R., Enghild, J.J., Martin, M.E., Jou, Y.-S., Myers, R.M., Roses, A.D., Vance, J.M., and Strittmatter, W.J. 1996. Huntingtin and DRPLA proteins selectively interact with the protein GAPDH. Nat. Med. 2 347–350. [DOI] [PubMed] [Google Scholar]
- Cooper, A.J.L., Sheu, K.-F.R., Burke, J.R., Onodera, O., Strittmatter, W.J., Roses, A.D., and Blass, J.P. 1997a. Polyglutamine domain are substrates of tissue transglutaminase. Does transglutaminase play a role in the expanded CAG/poly-Q neurodegenerative diseases? J. Neurochem. 69 431–434. [DOI] [PubMed] [Google Scholar]
- Cooper, A.J.L., Sheu, K.-F.R., Burke, J.R., Onodera, O., Strittmatter, W.J., Roses, A.D., and Blass, J.P. 1997b. Transglutaminase-catalysed inactivation of glyceraldehyde 3-phosphate dehydrogenase and a-ketoglutarate dehydrogenase complex by polyglutamine domains of pathological length. Proc. Natl. Acad. Sci. 94 12604–12609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper, A.J.L., Sheu, K.-F.R, Burke, J.R., Strittmatter, W.J., Gentile, V., Peluso, G., and Blass, J.P. 1999. Pathogenesis of inclusion bodies in (CAG)n/Qn-expansion diseases with special reference to the role of tissue transglutaminase and to selective vulnerability. J. Neurochem. 72 889–899. [DOI] [PubMed] [Google Scholar]
- Cooper, A.J.L., Wang, J., Pasternack, R., Fuchsbauer, H.L., Sheu, R.K., and Blass, J.P. 2000. Lysine-rich histone (H1) is a lysyl substrate of tissue transglutaminase: Possible involvement of transglutaminase in the formation of nuclear aggregates in (CAG)(n)/Q(n) expansion diseases. Dev. Neurosci. 22 404–417. [DOI] [PubMed] [Google Scholar]
- Davidson, B.E., Sajgo, M., Noller, H.F., and Harris, J.L. 1967. Amino acid sequence of glyceraldehyde 3-phosphate dehydrogenase from lobster muscle. Nature 216 1181–1185. [DOI] [PubMed] [Google Scholar]
- Davies, S.W., Turmaine, M., Cozens, B.A., DiFiglia, M., Sharp, A.H., Ross, C.A., Scherzinger, E., Wanker, E.E., Mangiarini, L., and Bates, G.P. 1997. Formation of neuronal intranuclear inclusions underlies the neurological dysfunction in mice transgenic for the HD mutation. Cell 90 537–548. [DOI] [PubMed] [Google Scholar]
- de Cristofaro, T., Affiatati, A., Cariello, L., Avvedimento, E.V., and Varrone, S. 1999. The length of polyglutamine tract, the rate of degradation, and the transglutaminase activity influence the formation of intracellular aggregates. Biochem. Biophys. Res. Commun. 260 150–158. [DOI] [PubMed] [Google Scholar]
- DiFiglia, M., Sapp, E., Chase, K.O., Davies, S.W., Bates, G.P., Vonsattel, J.P., and Aronin, N. 1997. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science 277 1990–1993. [DOI] [PubMed] [Google Scholar]
- Esposito, C., Mancuso, F., Calignano, A., Di Pierro, P., Pucci, P., and Porta, R. 1995. Neurokinin receptor could be differentiated by their capacity to respond to the transglutaminase-synthesized γ-(glutamyl5)spermine derivatives of Substance P. J. Neurochem. 65 420–426. [DOI] [PubMed] [Google Scholar]
- Esposito, C., Pucci, P., Amoresano, A., Marino, G., Cozzolino, A., and Porta, R. 1996. Transglutaminase from rat coagulating gland secretion. J. Biol. Chem. 271 27416–27423. [DOI] [PubMed] [Google Scholar]
- Esposito, C., Mariniello, L., Cozzolino, A., Amoresano, A., Orrù, S., and Porta, R. 2001. Rat coagulating gland secretion contains a kinesin heavy chain-like protein acting as type IV transglutaminase substrate. Biochemistry 40 4966–4971. [DOI] [PubMed] [Google Scholar]
- Folk, J.E. and Finlayson, J.S. 1977. The ɛ-(γ-glutamyl)lysine crosslink and the catalytic role of transglutaminases. Adv. Protein Chem. 31 1–133. [DOI] [PubMed] [Google Scholar]
- Gentile, V., Sepe, C., Calvani, M., Melone, M.A.B., Cotrufo, R., Cooper, A.J.L., Blass, J.P., and Peluso, G. 1998. Tissue transglutaminase-catalysed formation of high-molecular-weight aggegates in vitro is favored with long polyglutamine domains: A possible mechanism contributing to CAG-triplet diseases. Arch. Biochem. Biophys. 352 314–321. [DOI] [PubMed] [Google Scholar]
- Green, H. 1993. Human genetic diseases due to codon reiteration: Relationship to an evolutionary mechanism. Cell 74 955–956. [DOI] [PubMed] [Google Scholar]
- Groenen, P.J.T.A., Smulders, R.H.P.H., Peters, R.F.R., Grootjans, J.J, Van Den Ijessel, P.R.L.A., Bloemendal, H., and de Jong, W.W. 1994. The amino-donor substrate specificity of tissue-type transglutaminase. Influence of amino acid residues flanking the amnino-donor lysine residue. Eur. J. Biochem. 220 795–799. [DOI] [PubMed] [Google Scholar]
- Grootjans, J.J., Groenen, P.J.T.A., and de Jong, W.W. 1995. Substrate requirements for transglutaminases. Influence of the amino acid residue preceding the amino donor lysine in a native protein. J. Biol. Chem. 270 22855–22858. [DOI] [PubMed] [Google Scholar]
- Gutekunst, C.A, Li, S.H., Yi, H., Mulroy, J.S., Kuemmerle, S., Jones, R., Rye, D., Ferrante, R.J., Hersch, S.M., and Li, X.J. 1999. Nuclear and neuropil aggregates in Huntington's disease: Relationship to neuropathology. Neuroscience 19 2522–2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, J.I. and Perham, R.N. 1968. Glyceraldehyde 3-phosphate dehydrogenase from pig muscle. Nature 219 1025–1028. [DOI] [PubMed] [Google Scholar]
- Heinz, F. and Kulbe, K.D. 1970. D-glyceraldehyde 3-phosphate dehydrogenase from liver, isolation and characterisation of the bovine liver enzyme. Hoppe-Seylers Z. Physiol. Chem. 351 249–262. [DOI] [PubMed] [Google Scholar]
- Ikura, K., Kita, K., Fujita, I., Hashimoto, H., and Kawabata, N. 1998. Identification of amino acceptor protein substrates of transglutaminase in liver extracts: Use of 5–(biotinamido)pentylamine as a probe. Arch. Biochem. Biophys. 356 280–286. [DOI] [PubMed] [Google Scholar]
- Kahlem, P., Terré, C., Green, H., and Djian, P. 1996. Peptides containing glutamine repeats as substrates for transglutaminase-catalyzed cross-linking: Relevance to diseases of the nervous system. Proc. Natl. Acad. Sci. 93 14580–14585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S.-Y., Grant, P., Lee, J.-H., Pant, H.C., and Stainert, P.M. 1999. Differential expression of multiple transglutaminases in human brain. J. Biol. Chem. 274 30715–30721. [DOI] [PubMed] [Google Scholar]
- Kosby, B., Matilla, T., Burright, E.N., Merry, D.E., Fischbeck, K.H., Orr, H.T., and Zoghbi, H.Y. 1996. Spinocerebellar ataxia type-1 and spinobulbar muscular atrophy gene products interact with glyceraldehyde 3-phosphate dehydrogenase. Hum. Mol. Genet. 5 1311–1318. [DOI] [PubMed] [Google Scholar]
- Kraulis, P.J. 1991. MOLSCRIPT: A program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24 946–950. [Google Scholar]
- Laemmli, U.K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227 680–685. [DOI] [PubMed] [Google Scholar]
- Lambert, J.M. and Perham, R.N. 1977. Folding domains and intramolecular ionic interactions of lysine residues in glyceraldehyde 3-phosphate dehydrogenase. Biochem. J. 61 49–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, K.N., Birckbichler, P.J., and Patterson, M.K. 1989. GTP hydrolysis by guinea pig liver transglutaminase. Biochem. Biophys. Res. Commun. 162 1370–1375. [DOI] [PubMed] [Google Scholar]
- Li, H., Li, S.H., Cheng, A.L., Mangiarini, L., Bates, G.P., and Li, X.J. 1999. Ultrastructural localization and progressive formation of neuropil aggregates in Huntington's disease transgenic mice. Hum. Mol. Genet. 8 1227–1236. [DOI] [PubMed] [Google Scholar]
- Liu, Y., Gotte, G., Libonati, M., and Eisenberg, D. 2001. A domain-swapped RNase A dimer with implications for amyloid formation. Nat. Struct. Biol. 8 211–214. [DOI] [PubMed] [Google Scholar]
- Lorand, L. and Conrad, S.M. 1984. Transglutaminases. Mol. Cell. Biochem. 58 9–35. [DOI] [PubMed] [Google Scholar]
- Loewy, A.G., Blodgett, J.K., Blasé, F.R., and May, M.H. 1997. Synthesis and use of a substrate for the detection of isopeptidase activity. Anal. Biochem. 246 111–117. [DOI] [PubMed] [Google Scholar]
- Mariniello, L., Esposito, C., Di Pierro, P., Cozzolino, A., Pucci, P., and Porta, R.1993. Human immunodeficiency virus transmembrane glycoprotein gp41 is an amino acceptor and donor substrate for transglutaminase in vitro. Eur. J. Biochem. 215 99–104. [DOI] [PubMed] [Google Scholar]
- Matilla, A., Kosby, B.T., Cummings, C.J., Orr, H.A.T., and Zoghbi, H.Y. 1997. The leucine-rich acidic nuclear protein interacts with ataxin-1. Nature 389 974–978. [DOI] [PubMed] [Google Scholar]
- Melino, G. and Piacentini, M. 1998. "Tissue" transglutaminase in cell death: A downstream or a multifunctional upstream effector? FEBS Lett. 430 59–63. [DOI] [PubMed] [Google Scholar]
- Mercer, W.D., Winn, S.I., and Watson, H.C. 1976. Twinning in crystals of human skeletal muscle D-glyceraldehyde-3-phosphate dehydrogenase. J. Mol. Biol. 104 277–283. [DOI] [PubMed] [Google Scholar]
- Paulson, H.L. 1999. Human Genetics '99: Trinucleotide repeats. Protein fate in neurodegenerative proteinopathies: Polyglutamine diseases join the (mis)fold. Am. J. Hum. Genet. 64 339–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulson, H.L., Perez, M.K, Trottier, Y., Trojanowski, J.Q, Subramony, S.H., Das, S.S., Vig, P., Mandel, J.L., Fischbeck, K.H., and Pittman, R.N. 1997. Intranuclear inclusions of expanded polyglutamine protein in spinocerebellar ataxia type 3. Neuron 19 333–344. [DOI] [PubMed] [Google Scholar]
- Perutz, M.F. 1999. Glutamine repeats and neurodegenerative diseases: Molecular aspects. Trends Biochem. Sci. 24 58–63. [DOI] [PubMed] [Google Scholar]
- Perutz, M.F, Johnson, T., Suzuki, M., and Finch, J.T. 1994. Glutamine repeats as polar zippers: Their possible role in inherited neurodegenerative diseases. Proc. Natl. Acad. Sci. 91 5355–5358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porta, R., Esposito, C., Metafora, S., Pucci, P., Malorni, A., and Marino, G. 1988. Substance P as a transglutaminase substrate: Identification of the reaction products by fast atom bombardment mass spectrometry. Anal. Biochem. 172 499–503 [DOI] [PubMed] [Google Scholar]
- Porta, R., Esposito, C., Metafora, S., Malori, A., Pucci, P., Siciliano, R., and Marino, G. 1991. Mass spectrometric identification of the amino donor and acceptor sites in a transglutaminase protein substrates secreted from the rat seminal vesicles. Biochemistry 30 3114–3120. [DOI] [PubMed] [Google Scholar]
- Pucci, P., Malorni, A., Marino, G., Metafora, S., Esposito, C., and Porta, R. 1988. β-Endorphin modification by transglutaminase in vitro: Identification by FAB/MS of glutamine-11 and lysine-29 as acyl donor and acceptor sites. Biochem. Biophys. Res. Commun. 154 737–740. [DOI] [PubMed] [Google Scholar]
- Saunders, P.A., Chalecka-Franaszek, E., and Chuang D.M. 1997. Subcellular distribution of glyceraldehyde 3-phosphate dehydrogenase in cerebellar granule cells undergoing cytosine arabinoside-induced apoptosis. J. Neurochem. 69 1820–1828. [DOI] [PubMed] [Google Scholar]
- Sawa, A., Khan, A.A., Hester, L.D., and Snyder, S.H. 1997. Glyceraldehyde 3-phosphate dehydrogenase: Nuclear translocation in neuronal and nonneuronal cell death. Proc. Natl. Acad. Sci. 90 549–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirover, M.A. 1999. New insights into an old protein: The functional diversity of mammalian glyceraldehyde-3-phosphate dehydrogenase. Biochem. Biophys. Acta 1432 159–184. [DOI] [PubMed] [Google Scholar]
- Steinert, P.M., Candi, E., Tarcsa, E., Marekov, L.N., Sette, M., Paci, M., Ciani, B., Guerrieri, P., and Melino, G. 1999. Transglutaminase crosslinking and structural studies of the human small proline rich protein. Cell Death Differ. 6 916–930. [DOI] [PubMed] [Google Scholar]
- Tukamoto, T., Nukina, N., Ide, K., and Kanazawa, I. 1997. Huntington's disease gene product, huntingtin, associates with microtubules in vitro. Brain Res. Mol. Brain Res. 51 8–14. [DOI] [PubMed] [Google Scholar]
- Walker, J.E., Carne, A.F., Runswick, M.J., Bridgen, J., and Harris, J.I. 1980. D-glyceraldehyde 3-phosphate dehydrogenase. Complete amino acid sequence of the enzyme from Bacillus stearothermophilus. Eur. J. Biochem. 108 549–565. [DOI] [PubMed] [Google Scholar]
- Yoneda, Y. 1997. How proteins are transported from cytoplasm to nucleus. J. Biochem. 121 811–817. [DOI] [PubMed] [Google Scholar]