Abstract
Diploid (2n = 2x = 24) Solanum species with endosperm balance number (EBN) = 1 are sexually isolated from diploid 2EBN species and both tetraploid (2n = 4x = 48, 4EBN) and haploid (2n = 2x = 24, 2EBN) S. tuberosum Group Tuberosum. To sexually overcome these crossing barriers in the diploid species S. commersonii (1EBN), the manipulation of the EBN was accomplished by scaling up and down ploidy levels. Triploid F1 hybrids between an in vitro-doubled clone of S. commersonii (2n = 4x = 48, 2EBN) and diploid 2EBN clones were successfully used in 3x × 4x crosses with S. tuberosum Group Tuberosum, resulting in pentaploid/near pentaploid BC1 progenies. This provided evidence of 2n (3x) egg formation in the triploid female parents. Two selected BC1 pentaploid hybrids were successfully backcrossed both as male and as female parents with S. tuberosum Group Tuberosum. The somatic chromosome number varied greatly among the resulting BC2 progenies, which included hyperaneuploids, but also a number (4.8%) of 48-chromosome plants. The introgression of S. commersonii genomes was confirmed by the presence of S. commersonii-specific randomly amplified polymorphic DNA markers in the BC2 population analyzed. The results clearly demonstrate the feasibility of germplasm introgression from sexually isolated diploid 1EBN species into the 4x (4EBN) gene pool of the cultivated potato using sexual hybridization. Based on the amount and type of genetic variation generated, cumbersomeness, general applicability, costs, and other factors, it would be interesting to compare the approach reported here with other in vitro or in vivo, direct or indirect, approaches previously reported.
Keywords: 2n gametes, randomly amplified polymorphic DNA
The cultivated potato Solanum tuberosum Group Tuberosum (Tbr) (2n = 4x = 48) is a vegetatively propagated species with tetrasomic inheritance and a high level of heterozygosity (1). Hence, several important characteristics are strongly dependent on intralocus interactions, and genotypes with multi-allelic loci are superior to those that are mono- or diallelic. Unfortunately, due to the close relationship between cultivars, the genetic basis of modern varieties is rather limited. By contrast, wild Solanum species, most of which are diploid (2n = 2x = 24), represent a significantly greater source of allelic diversity than that present in the standard cultivars. They also possess many valuable traits, including resistances to biotic and abiotic stresses and superior tuber quality (2).
To introgress genes from diploid species into the cultivated potato, Tbr-4x can be crossed directly with 2n gamete-producing species to synthesize a tetraploid progeny. Alternatively, Tbr-4x can be crossed with hybrids between wild species and Tbr haploids. This second approach is preferable to reduce the number of wild genomes present in the tetraploid progeny. However, crosses between certain 24-chromosome species and Tbr repeatedly fail due to strong sexual isolating mechanisms. The most common underlying barrier to interspecific hybridization in Solanum spp. is the failure of endosperm development (3). According to the endosperm balance number (EBN) hypothesis (4), each Solanum species has a specific EBN, determined experimentally from crosses with tester species (5), and all successful crosses require a 2:1 maternal to paternal EBN ratio in the hybrid endosperm. EBN varies independently of ploidy, and interspecific crossability was found to depend on EBN, not ploidy (4–7). A number of diploid species have been assigned an EBN of 1 (5) and hence they cannot be crossed with either Tbr haploids (2EBN) or other diploid 2EBN species.
Solanum represents a model system to study sexual isolation mechanisms and ways to overcome them. To achieve the latter goal, both in vitro and in vivo approaches have been suggested. Somatic hybridization has been applied successfully, and tetraploid or hexaploid hybrids have been obtained between several 1EBN diploid species, including S. commersonii, and 2EBN Tbr haploids or 4EBN varieties, respectively (8, 9). A combination of rescue pollinations and embryo rescue has been used to exploit the diploid 1EBN species S. chancayense (10) and S. brevidens, S. etuberosum, and S. fernandezianum (11). The development of the EBN hypothesis led to the suggestion that in vivo EBN manipulations might be an efficient, simple, and reproducible approach for exploiting isolated germplasm (7, 12, 13). Inter-EBN introgression by in vivo manipulations was used to transfer genes from diploid S. commersonii and S. brevidens to S. chacoense and S. gourlay (2n = 2x = 24, 2EBN) background (14, 15), demonstrating an indirect means to infuse 1EBN species into Tbr via “bridge” species. The EBN hypothesis also predicts that direct infusion of 1EBN-2x germplasm into 4EBN-4x germplasm is possible, e. g., by the breeding scheme reported in Fig. 1. S. commersonii (Cmm) and Tbr were used to test this hypothesis from two standpoints, reproducibility and evidence of genetic introgression and recovery of usable germplasm.
Figure 1.
Breeding scheme followed to introgress S. commersonii (Cmm) into S. tuberosum Group Tuberosum (Tbr-4x) gene pool. A tetraploid clone of Cmm PI 243503 (Cmm-4x) was produced by in vitro tissue culture of leaf explants (16). Phureja-Tuberosum haploid hybrids (Phu-Tbr) came from the Wisconsin Breeding Program. Cmm-4x was crossed with Phu-Tbr, and resulting 3x F1 hybrids were backcrossed to Tbr-4x (cultivars Blondie, Carmine, Tollocan and clone Wis 482) to obtain BC1 5x hybrids. These were used as both female and as male parents in further backcrosses with Tbr-4x to produce PTH BC2 hybrids. 3x hybrids were coded as A1, B3, B10, B16, and C1; 5x hybrids were coded as P3 (B3 × Tbr-4x) and P5 (B10 × Tbr-4x).
MATERIALS AND METHODS
The triploid F1 hybrids were produced from 4x × 2x crosses between a tetraploid clone of Cmm PI 243503 (Cmm-4x) derived from in vitro tissue culture of leaf explants (16) and various Phureja-Tuberosum-haploid hybrids (Phu-Tbr) (17). Varieties Blondie, Carmine, and Tollocan as well as the advanced selection Wis 482 from the University of Wisconsin Breeding Program were used as tetraploid parents for their high male and female fertility.
Fig. 1 illustrates the crosses performed. Five triploid F1 hybrids (A1, B3, B10, B16, and C1) were used as female parents in 3x × 4x crosses with Tbr-4x. Pollinations were carried out in the greenhouse using emasculated flowers. Resulting seeds were treated with gibberellic acid (1,500 ppm) for 24 hr to break dormancy and sown in Styrofoam trays, and the seedlings obtained were transplanted to pots. Root tips were collected for mitotic analysis to check the ploidy level. Based on profuseness of flowering, vigor, and male fertility (evaluated as pollen stainability with 1% acetocarmine), two BC1 pentaploid clones obtained from 3x × Tbr-4x crosses (P3 and P5) were used both as male and as female parents in 5x × Tbr-4x and Tbr-4x × 5x crosses with varieties Blondie, Carmine, and Tollocan. The seeds were treated with gibberellic acid, sterilized with sodium hypochlorite (10% for 20 min), rinsed several times with sterilized water, and then sown in Petri dishes with Murashige and Skoog salts (18), sucrose 1%, and agar 0.8%. To vegetatively increase the genotypes, nodes with an axillary bud were excised from each of them and cultured in Magenta GA7 vessels supplemented with the same medium, at 50 μmol m−2 s−1, 16 hr light, 24°C. The chromosome number of a sample was checked by mitotic analysis of in vitro-grown roots as described (19).
To carry out randomly amplified polymorphic DNA (RAPD) analysis, DNA was extracted from leaf tissue by a modified hexadecyltrimethylammonium bromide (CTAB) procedure (19). RAPD markers were amplified by using 10-mer oligonucleotide primers and template DNA from Cmm (PI 243503), Phu-Tbr, B10, P5, and Tbr-4x, and from 17 genotypes from 5x × 4x crosses. A total of 64 oligonucleotides were examined, most of which were from the commercially available primer kits AN, AR, and H (Operon Technologies, Alameda, CA). The reaction conditions were as described by Williams et al. (20) using approximately 50 ng of DNA as template in a 25-μl reaction volume. Amplification was performed in a Perkin–Elmer/Cetus DNA thermal cycler programmed for 45 cycles of 1 min at 94°C, 1 min at 35°C, 2 min at 72°C, followed by 7 min at 72°C. Reaction products were resolved by electrophoresis in 1.5% agarose gels in 1× Tris-acetate-EDTA buffer.
RESULTS
Four of five 3x × Tbr-4x cross combinations produced berries with seeds after 20–40 pollinations per combination (Table 1). Berry set ranged from 39.1 (B3 × Tbr-4x) to 57.6 (B10 × Tbr-4x), but of 55 berries obtained, only 13 contained 1–2 seeds, the others being parthenocarpic. All derived plants were vigorous and tuberized under long day conditions. Most of them flowered profusely and shed stainable pollen. Mitotic analysis of root tip chromosomes indicated that most plants were pentaploid, rather than aneuploids.
Table 1.
Berry set, and average number of seeds/pollination and of seeds/berry from 3x × Tbr-4x crosses involving triploid (2n = 3x = 36) F1 hybrids and tetraploid (2n = 4x × 48) genotypes
| Triploid female parent* | Pollinations no. | Berries no.† | Berry set % | Average no. of seeds per
|
|
|---|---|---|---|---|---|
| Pollination | Berry‡ | ||||
| A1 | 29 | 13 (3) | 44.8 | 0.1 | 1.3 |
| B3 | 23 | 9 (2) | 39.1 | 0.2 | 2.5 |
| B10 | 26 | 15 (4) | 57.6 | 0.2 | 1.2 |
| B16 | 37 | 0 | — | — | — |
| C1 | 35 | 18 (4) | 51.4 | 0.1 | 1.2 |
| Average | 30 | 11 (3) | 36.7 | 0.1 | 1.5 |
Each 3x female parent was crossed with four different 4x male parents (varieties Tollocan, Blondie, Carmine, and clone Wis 482). No significant differences were found between crosses involving the same 3x female parent, so results were pooled across Tbr pollen parents.
The number of berries with seeds is reported in parentheses.
Only fruits with seeds were considered.
Two pentaploids were selected for further crosses with three cultivated varieties. The results from pooled 5x × Tbr-4x and Tbr-4x × 5x crosses are given in Table 2. The pentaploid genotypes could be easily crossed with the tetraploid varieties both as staminate and as pistillate parents, but 5x × Tbr-4x crosses gave better results than Tbr-4x × 5x crosses both in terms of berry set (47.7 vs. 28.5, respectively) and average number of seeds per berry (27.2 vs. 9.5, respectively). For comparison, results from Tbr-4x × Tbr-4x performed concurrently also are reported. Although berry set was similar to that of 5x × Tbr-4x crosses (48.2 vs. 47.7), the average number of seeds/berry (111.6) was much higher than that obtained from 5x-Tbr-4x crosses (27.2).
Table 2.
Berry set and average number of seeds/berry and of seeds/pollination from 5x × Tbr-4x and Tbr-4x × 5x crosses involving two pentaploid (2n = 5x = 60) BC1 hybrids (P5 and P3) and three tetraploid (2n = 4x = 48) varieties (Blondie, Carmine, and Tollocan)
| Cross combination* | Pollinations no. | Berries no. | Berry set % | Average no. of seeds per
|
|
|---|---|---|---|---|---|
| Pollination | Berry | ||||
| 5x × Tbr-4x | 58 | 30 | 47.7 | 17.2 | 27.2 |
| Tbr-4x × 5x | 44 | 16 | 28.5 | 1.9 | 9.5 |
| Tbr-4x × Tbr-4x† | 56 | 27 | 48.2 | 53.8 | 111.6 |
For comparisons, results from Tbr-4x × Tbr-4x crosses performed in the same environmental conditions also are given.
No significant differences were found between crosses within each cross combination, so results are pooled for cross combination.
Composite of crosses involving varieties Blondie, Carmine, and Tollocan.
The somatic chromosome numbers of BC2 progenies from crosses involving the pentaploid clones P3 and P5 and Tbr-4x were determined in a sample of plants (Table 3). The number of chromosomes observed among BC2 plants was highly varied, ranging from 48 to 60. Lower somatic chromosome numbers were predominant, manifesting a trend toward tetraploidy. Indeed, 44 analyzed plants (69.8%) had chromosome numbers ≤53. The morphology of these hybrids was varied, but their appearance resembled that of Tbr both in growth habit and tuber characteristics, and they crossed relatively easily with tetraploid potatoes.
Table 3.
Somatic chromosome number of BC2 genotypes from 5x × Tbr-4x and Tbr-4x × 5x crosses involving two pentaploid (2n = 5x = 60) BC1 hybrids (P3 and P5) and three tetraploid (2n = 4x = 48) varieties (Blondie, Carmine, and Tollocan)
| Cross combination | Genotypes analyzed, no. (%) | Genotypes with 48, 49, etc. somatic chromosomes, no. (%)
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 48 | 49 | 50 | 51 | 52 | 53 | 54 | 55 | 56 | 57 | 58 | 59 | 60 | ||
| P5 × Tbr-4x | 32 | 3 | 3 | 6 | 2 | 4 | 7 | 5 | 2 | 0 | 0 | 0 | 0 | 0 |
| P3 × Tbr-4x | 12 | 0 | 0 | 0 | 2 | 1 | 2 | 4 | 1 | 1 | 0 | 0 | 0 | 1 |
| 5x × Tbr-4x | 44 | 3 | 3 | 6 | 4 | 5 | 9 | 9 | 3 | 1 | 0 | 0 | 0 | 1 |
| (100) | (6.8) | (6.8) | (13.6) | (9.1) | (11.5) | (20.4) | (20.4) | (6.8) | (2.3) | (0) | (0) | (0) | (2.3) | |
| Tbr-4x × P5 | 10 | 0 | 1 | 1 | 2 | 2 | 3 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| Tbr-4x × P3 | 9 | 0 | 0 | 0 | 0 | 2 | 3 | 1 | 0 | 2 | 0 | 0 | 0 | 1 |
| Tbr-4x × 5x | 19 | 0 | 1 | 1 | 2 | 4 | 6 | 2 | 0 | 2 | 0 | 0 | 0 | 1 |
| (100) | (0) | (5.3) | (5.3) | (10.5) | (21.0) | (31.6) | (10.5) | (0) | (10.5) | (0) | (0) | (0) | (5.2) | |
| Total | 63 | 3 | 4 | 7 | 6 | 9 | 15 | 11 | 3 | 3 | 0 | 0 | 0 | 2 |
| (100) | (4.8) | (6.3) | (11.1) | (9.5) | (14.3) | (23.8) | (17.5) | (4.8) | (4.8) | (0) | (0) | (0) | (3.2) | |
Sixty-four decameric primers were screened to identify Cmm-specific RAPD markers. For 19 primers (29.7%) at least one amplified fragment was present in Cmm, in the 3x F1 hybrid B10 and 5x BC1 hybrid P5, but absent in Tbr-4x and in Phu-Tbr hybrids. These fragments were considered Cmm-specific RAPD markers and therefore were used to characterize 17 tetra- and hyper-tetraploid BC2 clones from 5x × 4x crosses involving the pentaploid hybrid P5 and different Tbr-4x varieties (Fig. 2). Table 4 reports the number of Cmm-specific RAPDs transmitted from the 5x hybrid P5 to the 5x × Tbr-4x hybrids. The number of Cmm-specific RAPDs present in the 5x × Tbr-4x hybrids ranged from 8 in PTH-4 (2n = 50) to 18 in PTH-3 (2n = 53), whereas in most of the genotypes (11 of 17) it was between 13 and 15. No significant correlation was found between the number of Cmm-specific RAPDs observed in the hybrids and their chromosome number (r = 0.312, P > 0.05).
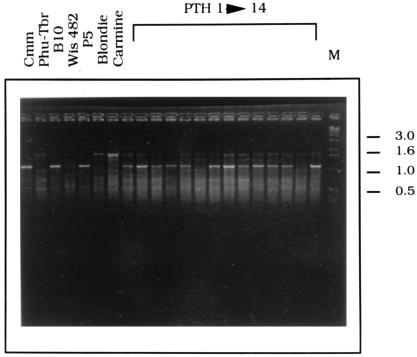
Figure 2.
RAPD marker 34 (TCGTAGCCAT) detecting one S. commersonii (Cmm)-specific band. DNA was amplified from Cmm, one Phureja-Tuberosum haploid hybrid (Phu-Tbr), one triploid (B10), one pentaploid (P5), three Tuberosum genotypes (Wis 482, Blondie, and Carmine), and 14 5x × Tbr-4x hybrids (PTH). M, molecular weight marker (kb).
Table 4.
Number of S. commersonii-specific loci observed in parental genotypes and in tetra- and hyper-tetraploid BC2 genotypes coming from 5x × Tbr-4x crosses as estimated on the basis of RAPD analysis
| Genotype* | Somatic chromosome number | Cmm-specific loci observed,† no. (%) |
|---|---|---|
| Parental genotypes | ||
| Cmm-2x | 24 | 19 (100) |
| Phu-Tbr | 24 | 0 (0) |
| B10 | 36 | 19 (100) |
| P5 | 60 | 19 (100) |
| Tbr-4x | 48 | 0 (0) |
| 5x × Tbr-4x hybrids | ||
| PTH-1 | 49 | 13 (68) |
| PTH-2 | 48 | 13 (68) |
| PTH-3 | 53 | 18 (95) |
| PTH-4 | 50 | 8 (42) |
| PTH-5 | 54 | 17 (89) |
| PTH-6 | 48 | 14 (74) |
| PTH-7 | 53 | 15 (79) |
| PTH-8 | 51 | 18 (95) |
| PTH-9 | 52 | 15 (79) |
| PTH-10 | 53 | 14 (74) |
| PTH-11 | 51 | 13 (68) |
| PTH-12 | 55 | 12 (63) |
| PTH-13 | 54 | 15 (79) |
| PTH-14 | 51 | 13 (68) |
| PTH-15 | 54 | 13 (68) |
| PTH-16 | 53 | 16 (84) |
| PTH-17 | 54 | 15 (79) |
Codes as described in Fig. 1.
19 Cmm-specific RAPDs were analyzed in each genotype. The percentage reported was calculated as follows: (no. of Cmm-specific RAPDs observed in each genotype/total no. of Cmm-specific RAPDs analyzed) × 100.
DISCUSSION
Solanum provides a model system where strong sexual isolating mechanisms exist, mainly due to “effective” ploidy (EBN). In this study, knowledge of EBN was used to manipulate whole sets of chromosomes for in vivo direct germplasm transfer from 2x (1EBN) Cmm to Tbr-4x. The first step was the use of a (2EBN) Cmm-4x clone obtained by in vitro regeneration from leaf explants of a diploid Cmm (16). This doubling technique already has been used with other Solanum species and exploits the somaclonal variation induced by the tissue culture conditions (21). The use of colchicine on meristems is less preferable due to drug toxicity and chimera formation.
The triploid hybrids obtained from 4x × 2x crosses have two haploid genomes of Cmm and one of Phu-Tbr. Hence, they have an EBN of 2 and can be crossed with tetraploid varieties (4EBN) only if they form 2n gametes. Indeed, 2n gametes from 3x plants are balanced functional gametes that lead to a compatible EBN ratio (2:1) in the hybrid endosperm. Novy and Hanneman (22) reported the production of triploids between Cmm and Tbr to direct transfer Cmm germplasm to Tbr, but due to the lack of fertile gametes, they never could be used in crosses with tetraploid varieties. The triploid F1 hybrids we produced flowered profusely and had a high pollen stainability, but they did not produce 2n pollen (17). On the other hand, they served as female parents in 3x × Tbr-4x crosses and gave rise to pentaploid/near pentaploid progenies, providing evidence of fertilization of 2n egg cells (3x) of the triploid parent by n (2x) sperm cells of the cultivated varieties. The production of near-pentaploid hybrids probably is due to the loss of chromosomes during megasporogenesis in the 3x female parent, perhaps by occasional omission from a restitution nucleus. The production of 5x BC1 progenies also confirmed the importance of 2n gametes in polyploidization and gene flow during evolution. If 4x-Cmm produced 2n eggs, it could be crossed with Tbr-4x to obtain 4EBN hexaploids. However, through hexaploids it is harder to restore the 48-chromosome number and pairing and gene exchange between nonhomologous chromosomes is not enforced, thus possibly limiting introgressive gene flow from wild species into the cultivated gene pool via recombination.
Ehlenfeldt and Hanneman (23) and Masuelli et al. (15) used 2x-2EBN bridge species (S. chacoense and S. gourlay, respectively) for indirect transfer of Cmm to Tbr. Indirect germplasm transfer through bridge species also has been accomplished by Adiwilaga and Brown (24) in the 4x-2EBN species S. acaule. Indirect transfer is less effective when compared with direct transfer, in that it has limited genetic efficiency due to the higher percentage of unadapted germplasm transmitted. In addition, the breeding program followed is longer and more time consuming due to the number of bridge crosses and backcrosses involved.
The pentaploid hybrids have two haploid Cmm genomes (1EBN) and three Tbr genomes (3EBN). As a result, their EBN is four, indicating no endosperm barriers to the introgression of genes from Cmm. Two pentaploid hybrids were backcrossed successfully both as female and as male parents with Tbr. In terms of berry and seed set they performed better as female than as male parents, presumably because chromosome imbalance is better tolerated in female than male gametophyte. In any case, the possibility of using pentaploids both as male and female parents will allow derivation of progenies with either Cmm or Tbr cytoplasm.
Progenies from 5x × Tbr-4x crosses had a trend toward tetraploidy, indicating that compared with the 5x parent a number of chromosomes had been lost. However, the presence of Cmm-specific RAPD markers in the BC2 population confirmed the introgression of Cmm genome. The high numbers of Cmm-RAPDs observed in 5x × Tbr-4x hybrids may suggest that several Cmm chromosomes or chromosome segments are still present in these plants. Despite the prevalence of Cmm markers, hybrids closely resembled Tbr in growth habit and tuber characteristics. Two possible explanations are the fact that markers lie outside highly expressed chromosomal regions (or at least those most critical to growth and tuberization) and the general dominance of the cultivated phenotype over the wild one.
Williams et al. (25) hypothesized bivalent pairing between homologous chromosomes to explain the large number of specific S. brevidens restriction fragment length polymorphism markers found in a pentaploid progeny between a hexaploid S. tuberosum (+) S. brevidens somatic hybrid, and S. tuberosum. The high percentage of Cmm-specific markers exhibited by most of the 5x × Tbr-4x hybrids and the preliminary diakinesis analysis of pentaploids (data not shown) may suggest that also in our study pairing was preferentially between the two Cmm-derived chromosome sets. Pairing behavior in our Cmm-Tbr sexual hybrids will be better understood with the use of mapped codominant markers and in situ hybridization technique.
Rapid progress in obtaining a large number of 48-chromosome genotypes is expected after one or two further backcross generations, and the availability of Cmm-specific RAPDs could speed up the selection of tetraploid Cmm-Tbr hybrids. The in vivo direct method we used also could be applied to exploit sexually isolated relatives of other species where an EBN system is operating. If the genetic recombination is limited, whole chromosomes can be moved across species to obtain addition or substitution lines.
Acknowledgments
This paper is contribution no. 150 from the Consiglio Nazionale delle Ricerche, Research Institute for Vegetable and Ornamental Plant Breeding.
ABBREVIATIONS
- EBN
endosperm balance number
- RAPD
randomly amplified polymorphic DNA
References
- 1.Howard H W. Genetics of Potato Solanum tuberosum. New York: Springer; 1970. p. 126. [Google Scholar]
- 2.Peloquin S J, Ortiz R. In: Plant Breeding in the 1990s. Stalker H T, Murphy J P, editors. Wallingford, U.K.: CAB International; 1991. pp. 485–507. [Google Scholar]
- 3.Hermsen J G, Th G. In: Potato Genetics. Bradshaw J E, Mackay G R, editors. Wallingford, U.K.: CAB International; 1994. pp. 515–538. [Google Scholar]
- 4.Johnston S A, den Nijs T M, Peloquin S J, Hanneman R E., Jr Theor Appl Genet. 1980;57:5–9. doi: 10.1007/BF00276002. [DOI] [PubMed] [Google Scholar]
- 5.Hanneman R E., Jr Euphytica. 1994;74:19–25. [Google Scholar]
- 6.Johnston S A, Hanneman R E., Jr Am Potato J. 1980;57:7–14. [Google Scholar]
- 7.Johnston S A, Hanneman R E., Jr Science. 1982;217:446–448. doi: 10.1126/science.217.4558.446. [DOI] [PubMed] [Google Scholar]
- 8.Cardi T, D’Ambrosio F, Consoli D, Puite K J, Ramulu K S. Theor Appl Genet. 1993;87:193–200. doi: 10.1007/BF00223764. [DOI] [PubMed] [Google Scholar]
- 9.Waara S, Glimelius K. Euphytica. 1995;85:217–233. [Google Scholar]
- 10.Singsit C, Hanneman R E., Jr Plant Cell Rep. 1991;9:475–478. doi: 10.1007/BF00232099. [DOI] [PubMed] [Google Scholar]
- 11.Watanabe K N, Orrillo M, Vega S, Valkonen J P T, Pehu E, Hurtado A, Tanksley SD. Genome. 1995;38:27–35. doi: 10.1139/g95-004. [DOI] [PubMed] [Google Scholar]
- 12.Ehlenfeldt M K, Hanneman R E., Jr Theor Appl Genet. 1988;75:825–832. [Google Scholar]
- 13.Ortiz R, Ehlenfeldt M K. Euphytica. 1992;60:105–113. [Google Scholar]
- 14.Peloquin S J, Jansky S H, Yerk G L. Am Potato J. 1989;66:629–638. [Google Scholar]
- 15.Masuelli R W, Camadro E L, Mendiburu A O. Genome. 1992;35:864–869. [Google Scholar]
- 16.Cardi T, Iannamico V, D’Ambrosio F, Filippone E, Lurquin P F. Plant Cell Tiss Org Cult. 1993;34:107–114. [Google Scholar]
- 17.Carputo D, Cardi T, Frusciante L, Peloquin S J. Euphytica. 1995;83:123–129. [Google Scholar]
- 18.Murashige T, Skoog F. Physiol Plant. 1962;15:473–497. [Google Scholar]
- 19.Bernatzky R, Tanksley S D. Plant Mol Biol Rep. 1986;4:37–41. [Google Scholar]
- 20.Williams J G K, Kubelik A R K, Livak J, Rafalski J A, Tingey S V. Nucleic Acids Res. 1990;18:6531–6535. doi: 10.1093/nar/18.22.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karp A, Risiott R, Jones M J K, Bright S W J. Plant Cell Tiss Org Cult. 1984;3:363–373. [Google Scholar]
- 22.Novy R G, Hanneman R E., Jr Am Potato J. 1991;68:151–169. [Google Scholar]
- 23.Ehlenfeldt M K, Hanneman R E., Jr Euphytica. 1988;37:181–187. [Google Scholar]
- 24.Adiwilaga K D, Brown C R. Theor Appl Genet. 1991;81:645–652. doi: 10.1007/BF00226732. [DOI] [PubMed] [Google Scholar]
- 25.Williams C E, Wielgus S M, Haberlach G T, Guenther C, Kim-Lee H, Helgeson J P. Genetics. 1993;135:1167–1173. doi: 10.1093/genetics/135.4.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]