Abstract
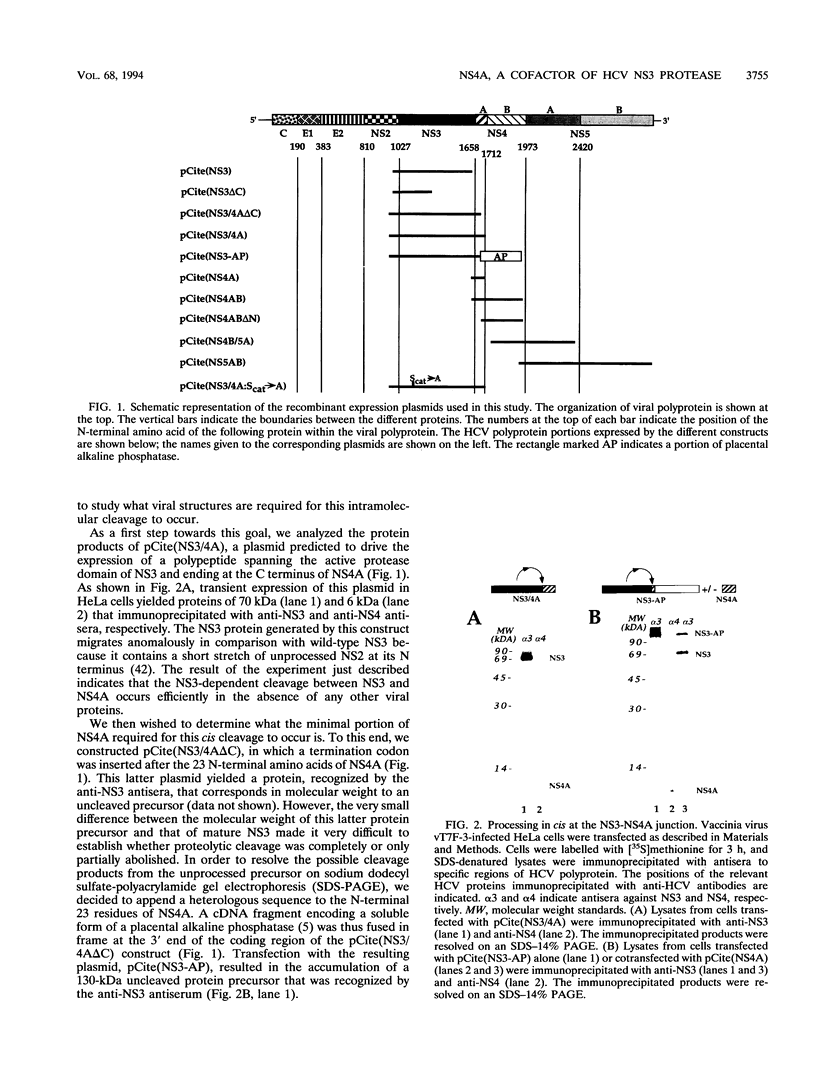
The proteolytic cleavages at the NS3-NS4A, NS4A-NS4B, NS4B-NS5A, and NS5A-NS5B junctions of hepatitis C virus (HCV) polyprotein are effected by the virus-encoded serine protease contained within NS3. Using transient expression in HeLa cells of cDNA fragments that code for regions of the HCV polyprotein, we studied whether viral functions other than NS3 are required for proteolytic processing at these sites. We found that, in addition to NS3, a C-terminal 33-amino-acid sequence of the NS4A protein is required for cleavage at the NS3-NS4A and NS4B-NS5A sites and that it accelerates the rate of cleavage at the NS5A-NS5B junction. In addition, we show that NS4A can activate the NS3 protease when supplied in trans. Our data suggest that HCV NS4A may be the functional analog of flavivirus NS2B and pestivirus p10 proteins.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arias C. F., Preugschat F., Strauss J. H. Dengue 2 virus NS2B and NS3 form a stable complex that can cleave NS3 within the helicase domain. Virology. 1993 Apr;193(2):888–899. doi: 10.1006/viro.1993.1198. [DOI] [PubMed] [Google Scholar]
- Baker D., Sohl J. L., Agard D. A. A protein-folding reaction under kinetic control. Nature. 1992 Mar 19;356(6366):263–265. doi: 10.1038/356263a0. [DOI] [PubMed] [Google Scholar]
- Bartenschlager R., Ahlborn-Laake L., Mous J., Jacobsen H. Nonstructural protein 3 of the hepatitis C virus encodes a serine-type proteinase required for cleavage at the NS3/4 and NS4/5 junctions. J Virol. 1993 Jul;67(7):3835–3844. doi: 10.1128/jvi.67.7.3835-3844.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazan J. F., Fletterick R. J. Detection of a trypsin-like serine protease domain in flaviviruses and pestiviruses. Virology. 1989 Aug;171(2):637–639. doi: 10.1016/0042-6822(89)90639-9. [DOI] [PubMed] [Google Scholar]
- Berger J., Hauber J., Hauber R., Geiger R., Cullen B. R. Secreted placental alkaline phosphatase: a powerful new quantitative indicator of gene expression in eukaryotic cells. Gene. 1988 Jun 15;66(1):1–10. doi: 10.1016/0378-1119(88)90219-3. [DOI] [PubMed] [Google Scholar]
- Chambers T. J., Grakoui A., Rice C. M. Processing of the yellow fever virus nonstructural polyprotein: a catalytically active NS3 proteinase domain and NS2B are required for cleavages at dibasic sites. J Virol. 1991 Nov;65(11):6042–6050. doi: 10.1128/jvi.65.11.6042-6050.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers T. J., Hahn C. S., Galler R., Rice C. M. Flavivirus genome organization, expression, and replication. Annu Rev Microbiol. 1990;44:649–688. doi: 10.1146/annurev.mi.44.100190.003245. [DOI] [PubMed] [Google Scholar]
- Chambers T. J., Nestorowicz A., Amberg S. M., Rice C. M. Mutagenesis of the yellow fever virus NS2B protein: effects on proteolytic processing, NS2B-NS3 complex formation, and viral replication. J Virol. 1993 Nov;67(11):6797–6807. doi: 10.1128/jvi.67.11.6797-6807.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers T. J., Weir R. C., Grakoui A., McCourt D. W., Bazan J. F., Fletterick R. J., Rice C. M. Evidence that the N-terminal domain of nonstructural protein NS3 from yellow fever virus is a serine protease responsible for site-specific cleavages in the viral polyprotein. Proc Natl Acad Sci U S A. 1990 Nov;87(22):8898–8902. doi: 10.1073/pnas.87.22.8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien D. Y., Choo Q. L., Tabrizi A., Kuo C., McFarland J., Berger K., Lee C., Shuster J. R., Nguyen T., Moyer D. L. Diagnosis of hepatitis C virus (HCV) infection using an immunodominant chimeric polyprotein to capture circulating antibodies: reevaluation of the role of HCV in liver disease. Proc Natl Acad Sci U S A. 1992 Nov 1;89(21):10011–10015. doi: 10.1073/pnas.89.21.10011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo Q. L., Kuo G., Weiner A. J., Overby L. R., Bradley D. W., Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989 Apr 21;244(4902):359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- Choo Q. L., Richman K. H., Han J. H., Berger K., Lee C., Dong C., Gallegos C., Coit D., Medina-Selby R., Barr P. J. Genetic organization and diversity of the hepatitis C virus. Proc Natl Acad Sci U S A. 1991 Mar 15;88(6):2451–2455. doi: 10.1073/pnas.88.6.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collett M. S. Molecular genetics of pestiviruses. Comp Immunol Microbiol Infect Dis. 1992 Jul;15(3):145–154. doi: 10.1016/0147-9571(92)90087-8. [DOI] [PubMed] [Google Scholar]
- Craig E. A. Chaperones: helpers along the pathways to protein folding. Science. 1993 Jun 25;260(5116):1902–1903. doi: 10.1126/science.8100364. [DOI] [PubMed] [Google Scholar]
- Eckart M. R., Selby M., Masiarz F., Lee C., Berger K., Crawford K., Kuo C., Kuo G., Houghton M., Choo Q. L. The hepatitis C virus encodes a serine protease involved in processing of the putative nonstructural proteins from the viral polyprotein precursor. Biochem Biophys Res Commun. 1993 Apr 30;192(2):399–406. doi: 10.1006/bbrc.1993.1429. [DOI] [PubMed] [Google Scholar]
- Elroy-Stein O., Fuerst T. R., Moss B. Cap-independent translation of mRNA conferred by encephalomyocarditis virus 5' sequence improves the performance of the vaccinia virus/bacteriophage T7 hybrid expression system. Proc Natl Acad Sci U S A. 1989 Aug;86(16):6126–6130. doi: 10.1073/pnas.86.16.6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falgout B., Miller R. H., Lai C. J. Deletion analysis of dengue virus type 4 nonstructural protein NS2B: identification of a domain required for NS2B-NS3 protease activity. J Virol. 1993 Apr;67(4):2034–2042. doi: 10.1128/jvi.67.4.2034-2042.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falgout B., Pethel M., Zhang Y. M., Lai C. J. Both nonstructural proteins NS2B and NS3 are required for the proteolytic processing of dengue virus nonstructural proteins. J Virol. 1991 May;65(5):2467–2475. doi: 10.1128/jvi.65.5.2467-2475.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuerst T. R., Niles E. G., Studier F. W., Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci U S A. 1986 Nov;83(21):8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grakoui A., McCourt D. W., Wychowski C., Feinstone S. M., Rice C. M. A second hepatitis C virus-encoded proteinase. Proc Natl Acad Sci U S A. 1993 Nov 15;90(22):10583–10587. doi: 10.1073/pnas.90.22.10583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grakoui A., McCourt D. W., Wychowski C., Feinstone S. M., Rice C. M. Characterization of the hepatitis C virus-encoded serine proteinase: determination of proteinase-dependent polyprotein cleavage sites. J Virol. 1993 May;67(5):2832–2843. doi: 10.1128/jvi.67.5.2832-2843.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grakoui A., Wychowski C., Lin C., Feinstone S. M., Rice C. M. Expression and identification of hepatitis C virus polyprotein cleavage products. J Virol. 1993 Mar;67(3):1385–1395. doi: 10.1128/jvi.67.3.1385-1395.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hijikata M., Kato N., Ootsuyama Y., Nakagawa M., Shimotohno K. Gene mapping of the putative structural region of the hepatitis C virus genome by in vitro processing analysis. Proc Natl Acad Sci U S A. 1991 Jul 1;88(13):5547–5551. doi: 10.1073/pnas.88.13.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hijikata M., Mizushima H., Akagi T., Mori S., Kakiuchi N., Kato N., Tanaka T., Kimura K., Shimotohno K. Two distinct proteinase activities required for the processing of a putative nonstructural precursor protein of hepatitis C virus. J Virol. 1993 Aug;67(8):4665–4675. doi: 10.1128/jvi.67.8.4665-4675.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikemura H., Inouye M. In vitro processing of pro-subtilisin produced in Escherichia coli. J Biol Chem. 1988 Sep 15;263(26):12959–12963. [PubMed] [Google Scholar]
- Jang S. K., Davies M. V., Kaufman R. J., Wimmer E. Initiation of protein synthesis by internal entry of ribosomes into the 5' nontranslated region of encephalomyocarditis virus RNA in vivo. J Virol. 1989 Apr;63(4):1651–1660. doi: 10.1128/jvi.63.4.1651-1660.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jore J., De Geus B., Jackson R. J., Pouwels P. H., Enger-Valk B. E. Poliovirus protein 3CD is the active protease for processing of the precursor protein P1 in vitro. J Gen Virol. 1988 Jul;69(Pt 7):1627–1636. doi: 10.1099/0022-1317-69-7-1627. [DOI] [PubMed] [Google Scholar]
- Kato N., Hijikata M., Ootsuyama Y., Nakagawa M., Ohkoshi S., Sugimura T., Shimotohno K. Molecular cloning of the human hepatitis C virus genome from Japanese patients with non-A, non-B hepatitis. Proc Natl Acad Sci U S A. 1990 Dec;87(24):9524–9528. doi: 10.1073/pnas.87.24.9524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh I., Ikawa Y., Yoshinaka Y. Retrovirus protease characterized as a dimeric aspartic proteinase. J Virol. 1989 May;63(5):2226–2232. doi: 10.1128/jvi.63.5.2226-2232.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo G., Choo Q. L., Alter H. J., Gitnick G. L., Redeker A. G., Purcell R. H., Miyamura T., Dienstag J. L., Alter M. J., Stevens C. E. An assay for circulating antibodies to a major etiologic virus of human non-A, non-B hepatitis. Science. 1989 Apr 21;244(4902):362–364. doi: 10.1126/science.2496467. [DOI] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Lin C., Amberg S. M., Chambers T. J., Rice C. M. Cleavage at a novel site in the NS4A region by the yellow fever virus NS2B-3 proteinase is a prerequisite for processing at the downstream 4A/4B signalase site. J Virol. 1993 Apr;67(4):2327–2335. doi: 10.1128/jvi.67.4.2327-2335.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller R. H., Purcell R. H. Hepatitis C virus shares amino acid sequence similarity with pestiviruses and flaviviruses as well as members of two plant virus supergroups. Proc Natl Acad Sci U S A. 1990 Mar;87(6):2057–2061. doi: 10.1073/pnas.87.6.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizzi E., Tramontano A., Tomei L., La Monica N., Failla C., Sardana M., Wood T., De Francesco R. Molecular model of the specificity pocket of the hepatitis C virus protease: implications for substrate recognition. Proc Natl Acad Sci U S A. 1994 Feb 1;91(3):888–892. doi: 10.1073/pnas.91.3.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preugschat F., Yao C. W., Strauss J. H. In vitro processing of dengue virus type 2 nonstructural proteins NS2A, NS2B, and NS3. J Virol. 1990 Sep;64(9):4364–4374. doi: 10.1128/jvi.64.9.4364-4374.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santolini E., Migliaccio G., La Monica N. Biosynthesis and biochemical properties of the hepatitis C virus core protein. J Virol. 1994 Jun;68(6):3631–3641. doi: 10.1128/jvi.68.6.3631-3641.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silen J. L., Agard D. A. The alpha-lytic protease pro-region does not require a physical linkage to activate the protease domain in vivo. Nature. 1989 Oct 5;341(6241):462–464. doi: 10.1038/341462a0. [DOI] [PubMed] [Google Scholar]
- Takamizawa A., Mori C., Fuke I., Manabe S., Murakami S., Fujita J., Onishi E., Andoh T., Yoshida I., Okayama H. Structure and organization of the hepatitis C virus genome isolated from human carriers. J Virol. 1991 Mar;65(3):1105–1113. doi: 10.1128/jvi.65.3.1105-1113.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomei L., Failla C., Santolini E., De Francesco R., La Monica N. NS3 is a serine protease required for processing of hepatitis C virus polyprotein. J Virol. 1993 Jul;67(7):4017–4026. doi: 10.1128/jvi.67.7.4017-4026.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos P., Verver J., Jaegle M., Wellink J., van Kammen A., Goldbach R. Two viral proteins involved in the proteolytic processing of the cowpea mosaic virus polyproteins. Nucleic Acids Res. 1988 Mar 25;16(5):1967–1985. doi: 10.1093/nar/16.5.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster A., Hay R. T., Kemp G. The adenovirus protease is activated by a virus-coded disulphide-linked peptide. Cell. 1993 Jan 15;72(1):97–104. doi: 10.1016/0092-8674(93)90053-s. [DOI] [PubMed] [Google Scholar]
- Wiskerchen M., Collett M. S. Pestivirus gene expression: protein p80 of bovine viral diarrhea virus is a proteinase involved in polyprotein processing. Virology. 1991 Sep;184(1):341–350. doi: 10.1016/0042-6822(91)90850-b. [DOI] [PubMed] [Google Scholar]
- Ypma-Wong M. F., Dewalt P. G., Johnson V. H., Lamb J. G., Semler B. L. Protein 3CD is the major poliovirus proteinase responsible for cleavage of the P1 capsid precursor. Virology. 1988 Sep;166(1):265–270. doi: 10.1016/0042-6822(88)90172-9. [DOI] [PubMed] [Google Scholar]
- Zhu X. L., Ohta Y., Jordan F., Inouye M. Pro-sequence of subtilisin can guide the refolding of denatured subtilisin in an intermolecular process. Nature. 1989 Jun 8;339(6224):483–484. doi: 10.1038/339483a0. [DOI] [PubMed] [Google Scholar]