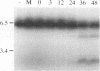
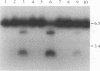
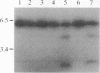
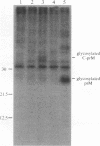
Abstract
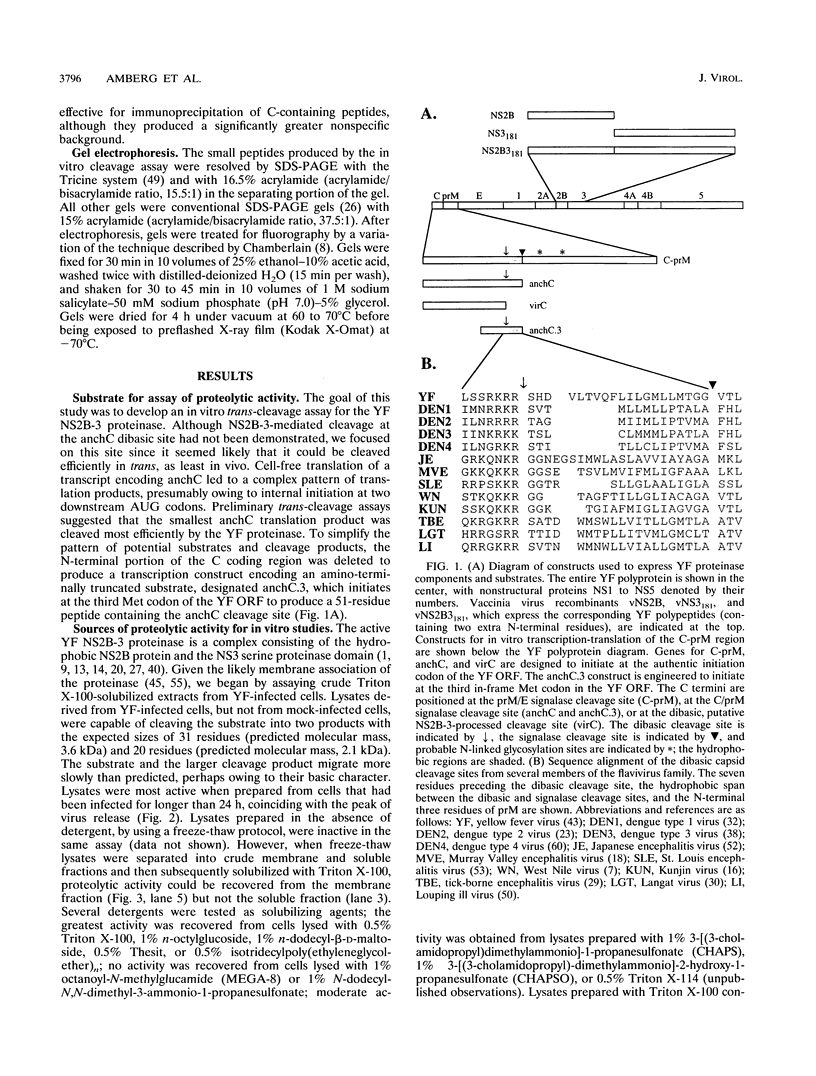
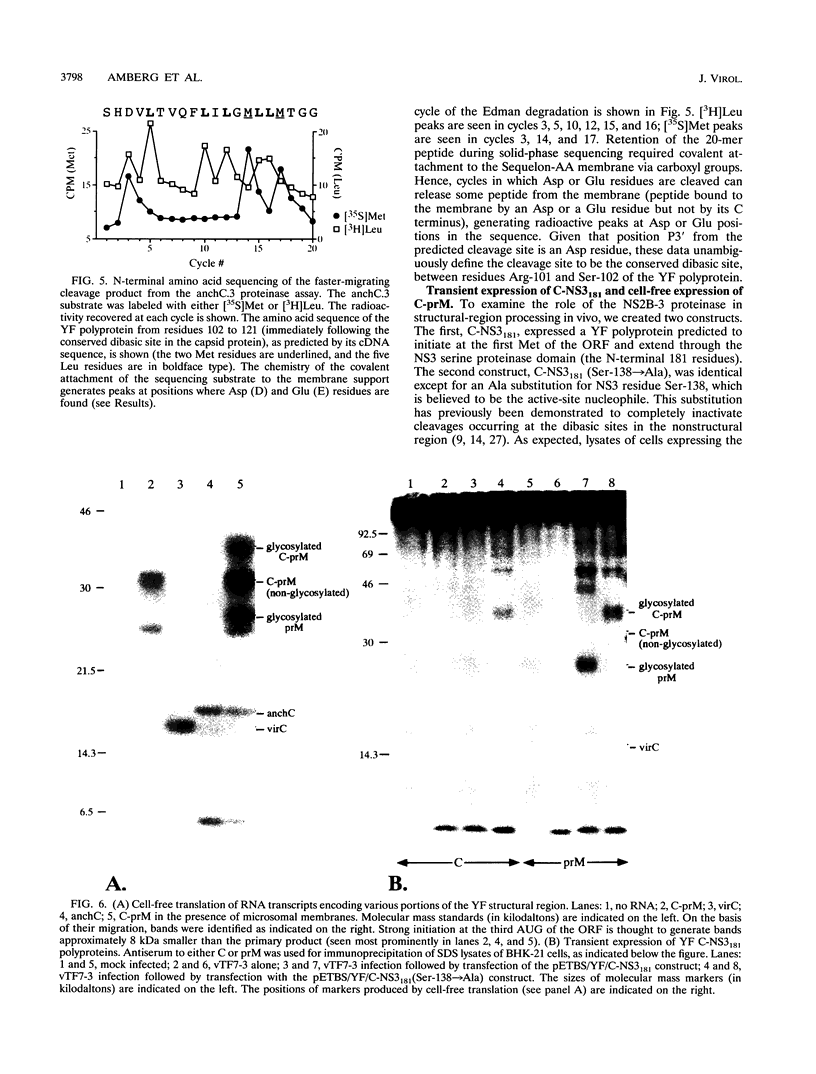
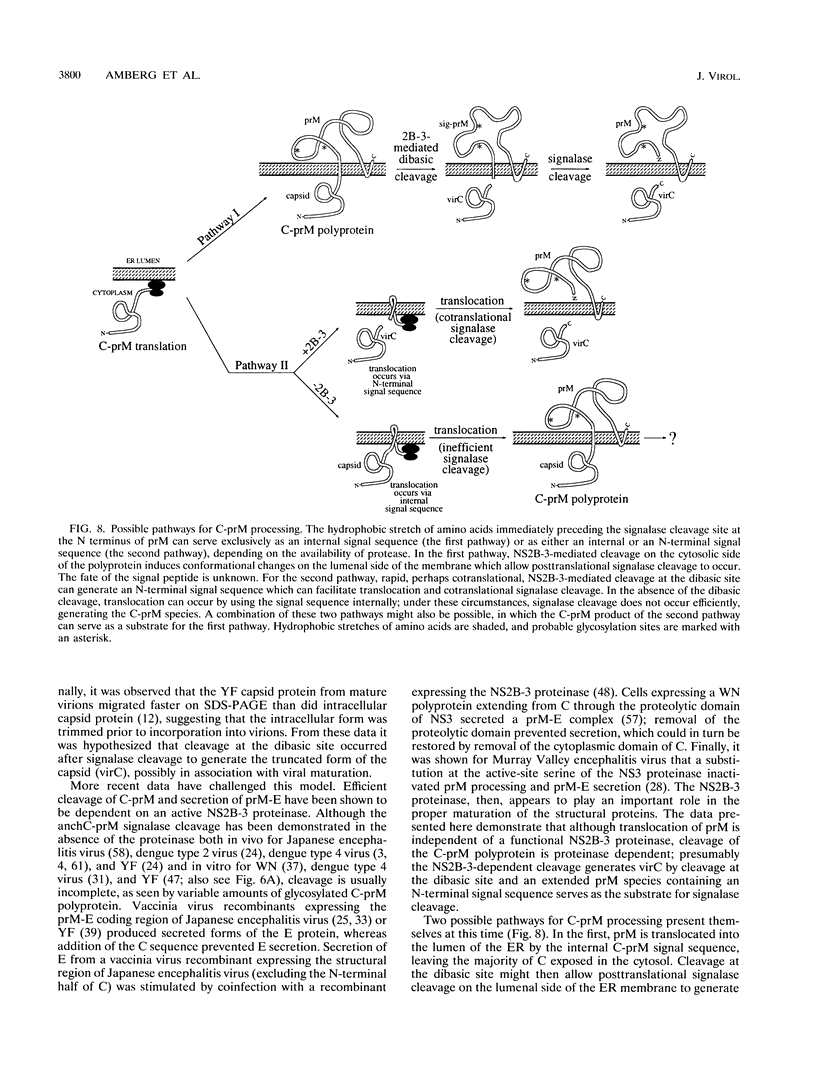
Several of the cleavages required to generate the mature nonstructural proteins from the flaviviral polyprotein are known to be mediated by a complex consisting of NS2B and a serine proteinase domain located in the N-terminal one-third of NS3. These cleavages typically occur after two basic residues followed by a short side chain residue. Cleavage at a similar dibasic site in the structural region is believed to produce the C terminus of the virion capsid protein. To study this cleavage, we developed a cell-free trans cleavage assay for yellow fever virus (YF)-specific proteolytic activity by using a substrate spanning the C protein dibasic site. Cleavage at the predicted site was observed when the substrate was incubated with detergent-solubilized lysates from YF-infected BHK cells. NS2B and the NS3 proteinase domain were the only YF-specific proteins required for this cleavage. Cell fractionation studies demonstrated that the YF-specific proteolytic activity was membrane associated and that activity could be detected only after detergent solubilization. Previous cell-free studies led to a hypothesis that processing in the C-prM region involves (i) translation of C followed by translocation and core glycosylation of prM by using an internal signal sequence, (ii) signalase cleavage to produce a membrane-anchored form of the C protein (anchC) and the N terminus of prM, and (iii) NS2B-3-mediated cleavage at the anchC dibasic site to produce the C terminus of the virion C protein. However, the results of in vivo transient-expression studies do not support this temporal cleavage order. Rather, expression of a YF polyprotein extending from C through the N-terminal one-third of NS3 revealed that C-prM processing, but not translocation, was dependent on an active NS2B-3 proteinase. This suggests that signalase-mediated cleavage in the lumen of the endoplasmic reticulum may be dependent on prior cleavage at the anchC dibasic site. Possible pathways for processing in the C-prM region are outlined and discussed.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arias C. F., Preugschat F., Strauss J. H. Dengue 2 virus NS2B and NS3 form a stable complex that can cleave NS3 within the helicase domain. Virology. 1993 Apr;193(2):888–899. doi: 10.1006/viro.1993.1198. [DOI] [PubMed] [Google Scholar]
- Bodwell J. E., Ortí E., Coull J. M., Pappin D. J., Smith L. I., Swift F. Identification of phosphorylated sites in the mouse glucocorticoid receptor. J Biol Chem. 1991 Apr 25;266(12):7549–7555. [PubMed] [Google Scholar]
- Bray M., Lai C. J. Dengue virus premembrane and membrane proteins elicit a protective immune response. Virology. 1991 Nov;185(1):505–508. doi: 10.1016/0042-6822(91)90809-p. [DOI] [PubMed] [Google Scholar]
- Bray M., Zhao B. T., Markoff L., Eckels K. H., Chanock R. M., Lai C. J. Mice immunized with recombinant vaccinia virus expressing dengue 4 virus structural proteins with or without nonstructural protein NS1 are protected against fatal dengue virus encephalitis. J Virol. 1989 Jun;63(6):2853–2856. doi: 10.1128/jvi.63.6.2853-2856.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahour A., Falgout B., Lai C. J. Cleavage of the dengue virus polyprotein at the NS3/NS4A and NS4B/NS5 junctions is mediated by viral protease NS2B-NS3, whereas NS4A/NS4B may be processed by a cellular protease. J Virol. 1992 Mar;66(3):1535–1542. doi: 10.1128/jvi.66.3.1535-1542.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castle E., Nowak T., Leidner U., Wengler G., Wengler G. Sequence analysis of the viral core protein and the membrane-associated proteins V1 and NV2 of the flavivirus West Nile virus and of the genome sequence for these proteins. Virology. 1985 Sep;145(2):227–236. doi: 10.1016/0042-6822(85)90156-4. [DOI] [PubMed] [Google Scholar]
- Chamberlain J. P. Fluorographic detection of radioactivity in polyacrylamide gels with the water-soluble fluor, sodium salicylate. Anal Biochem. 1979 Sep 15;98(1):132–135. doi: 10.1016/0003-2697(79)90716-4. [DOI] [PubMed] [Google Scholar]
- Chambers T. J., Grakoui A., Rice C. M. Processing of the yellow fever virus nonstructural polyprotein: a catalytically active NS3 proteinase domain and NS2B are required for cleavages at dibasic sites. J Virol. 1991 Nov;65(11):6042–6050. doi: 10.1128/jvi.65.11.6042-6050.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers T. J., Hahn C. S., Galler R., Rice C. M. Flavivirus genome organization, expression, and replication. Annu Rev Microbiol. 1990;44:649–688. doi: 10.1146/annurev.mi.44.100190.003245. [DOI] [PubMed] [Google Scholar]
- Chambers T. J., McCourt D. W., Rice C. M. Production of yellow fever virus proteins in infected cells: identification of discrete polyprotein species and analysis of cleavage kinetics using region-specific polyclonal antisera. Virology. 1990 Jul;177(1):159–174. doi: 10.1016/0042-6822(90)90470-c. [DOI] [PubMed] [Google Scholar]
- Chambers T. J., McCourt D. W., Rice C. M. Yellow fever virus proteins NS2A, NS2B, and NS4B: identification and partial N-terminal amino acid sequence analysis. Virology. 1989 Mar;169(1):100–109. doi: 10.1016/0042-6822(89)90045-7. [DOI] [PubMed] [Google Scholar]
- Chambers T. J., Nestorowicz A., Amberg S. M., Rice C. M. Mutagenesis of the yellow fever virus NS2B protein: effects on proteolytic processing, NS2B-NS3 complex formation, and viral replication. J Virol. 1993 Nov;67(11):6797–6807. doi: 10.1128/jvi.67.11.6797-6807.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers T. J., Weir R. C., Grakoui A., McCourt D. W., Bazan J. F., Fletterick R. J., Rice C. M. Evidence that the N-terminal domain of nonstructural protein NS3 from yellow fever virus is a serine protease responsible for site-specific cleavages in the viral polyprotein. Proc Natl Acad Sci U S A. 1990 Nov;87(22):8898–8902. doi: 10.1073/pnas.87.22.8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo Q. L., Kuo G., Weiner A. J., Overby L. R., Bradley D. W., Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989 Apr 21;244(4902):359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- Coia G., Parker M. D., Speight G., Byrne M. E., Westaway E. G. Nucleotide and complete amino acid sequences of Kunjin virus: definitive gene order and characteristics of the virus-specified proteins. J Gen Virol. 1988 Jan;69(Pt 1):1–21. doi: 10.1099/0022-1317-69-1-1. [DOI] [PubMed] [Google Scholar]
- Collett M. S., Anderson D. K., Retzel E. Comparisons of the pestivirus bovine viral diarrhoea virus with members of the flaviviridae. J Gen Virol. 1988 Oct;69(Pt 10):2637–2643. doi: 10.1099/0022-1317-69-10-2637. [DOI] [PubMed] [Google Scholar]
- Dalgarno L., Trent D. W., Strauss J. H., Rice C. M. Partial nucleotide sequence of the Murray Valley encephalitis virus genome. Comparison of the encoded polypeptides with yellow fever virus structural and non-structural proteins. J Mol Biol. 1986 Feb 5;187(3):309–323. doi: 10.1016/0022-2836(86)90435-3. [DOI] [PubMed] [Google Scholar]
- Falgout B., Chanock R., Lai C. J. Proper processing of dengue virus nonstructural glycoprotein NS1 requires the N-terminal hydrophobic signal sequence and the downstream nonstructural protein NS2a. J Virol. 1989 May;63(5):1852–1860. doi: 10.1128/jvi.63.5.1852-1860.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falgout B., Pethel M., Zhang Y. M., Lai C. J. Both nonstructural proteins NS2B and NS3 are required for the proteolytic processing of dengue virus nonstructural proteins. J Virol. 1991 May;65(5):2467–2475. doi: 10.1128/jvi.65.5.2467-2475.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuerst T. R., Niles E. G., Studier F. W., Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci U S A. 1986 Nov;83(21):8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn C. S., Hahn Y. S., Braciale T. J., Rice C. M. Infectious Sindbis virus transient expression vectors for studying antigen processing and presentation. Proc Natl Acad Sci U S A. 1992 Apr 1;89(7):2679–2683. doi: 10.1073/pnas.89.7.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn Y. S., Galler R., Hunkapiller T., Dalrymple J. M., Strauss J. H., Strauss E. G. Nucleotide sequence of dengue 2 RNA and comparison of the encoded proteins with those of other flaviviruses. Virology. 1988 Jan;162(1):167–180. doi: 10.1016/0042-6822(88)90406-0. [DOI] [PubMed] [Google Scholar]
- Hahn Y. S., Lenches E. M., Galler R., Rice C. M., Dalrymple J., Strauss J. H. Expression of the structural proteins of dengue 2 virus and yellow fever virus by recombinant vaccinia viruses. Arch Virol. 1990;115(3-4):251–265. doi: 10.1007/BF01310534. [DOI] [PubMed] [Google Scholar]
- Konishi E., Pincus S., Fonseca B. A., Shope R. E., Paoletti E., Mason P. W. Comparison of protective immunity elicited by recombinant vaccinia viruses that synthesize E or NS1 of Japanese encephalitis virus. Virology. 1991 Nov;185(1):401–410. doi: 10.1016/0042-6822(91)90788-d. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lin C., Amberg S. M., Chambers T. J., Rice C. M. Cleavage at a novel site in the NS4A region by the yellow fever virus NS2B-3 proteinase is a prerequisite for processing at the downstream 4A/4B signalase site. J Virol. 1993 Apr;67(4):2327–2335. doi: 10.1128/jvi.67.4.2327-2335.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobigs M. Flavivirus premembrane protein cleavage and spike heterodimer secretion require the function of the viral proteinase NS3. Proc Natl Acad Sci U S A. 1993 Jul 1;90(13):6218–6222. doi: 10.1073/pnas.90.13.6218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandl C. W., Heinz F. X., Kunz C. Sequence of the structural proteins of tick-borne encephalitis virus (western subtype) and comparative analysis with other flaviviruses. Virology. 1988 Sep;166(1):197–205. doi: 10.1016/0042-6822(88)90161-4. [DOI] [PubMed] [Google Scholar]
- Mandl C. W., Iacono-Connors L., Wallner G., Holzmann H., Kunz C., Heinz F. X. Sequence of the genes encoding the structural proteins of the low-virulence tick-borne flaviviruses Langat TP21 and Yelantsev. Virology. 1991 Dec;185(2):891–895. doi: 10.1016/0042-6822(91)90567-u. [DOI] [PubMed] [Google Scholar]
- Markoff L. In vitro processing of dengue virus structural proteins: cleavage of the pre-membrane protein. J Virol. 1989 Aug;63(8):3345–3352. doi: 10.1128/jvi.63.8.3345-3352.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason P. W., McAda P. C., Mason T. L., Fournier M. J. Sequence of the dengue-1 virus genome in the region encoding the three structural proteins and the major nonstructural protein NS1. Virology. 1987 Nov;161(1):262–267. doi: 10.1016/0042-6822(87)90196-6. [DOI] [PubMed] [Google Scholar]
- Mason P. W., Pincus S., Fournier M. J., Mason T. L., Shope R. E., Paoletti E. Japanese encephalitis virus-vaccinia recombinants produce particulate forms of the structural membrane proteins and induce high levels of protection against lethal JEV infection. Virology. 1991 Jan;180(1):294–305. doi: 10.1016/0042-6822(91)90034-9. [DOI] [PubMed] [Google Scholar]
- Matsudaira P. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J Biol Chem. 1987 Jul 25;262(21):10035–10038. [PubMed] [Google Scholar]
- Murray J. M., Aaskov J. G., Wright P. J. Processing of the dengue virus type 2 proteins prM and C-prM. J Gen Virol. 1993 Feb;74(Pt 2):175–182. doi: 10.1099/0022-1317-74-2-175. [DOI] [PubMed] [Google Scholar]
- Nestorowicz A., Chambers T. J., Rice C. M. Mutagenesis of the yellow fever virus NS2A/2B cleavage site: effects on proteolytic processing, viral replication, and evidence for alternative processing of the NS2A protein. Virology. 1994 Feb 15;199(1):114–123. doi: 10.1006/viro.1994.1103. [DOI] [PubMed] [Google Scholar]
- Nowak T., Färber P. M., Wengler G., Wengler G. Analyses of the terminal sequences of West Nile virus structural proteins and of the in vitro translation of these proteins allow the proposal of a complete scheme of the proteolytic cleavages involved in their synthesis. Virology. 1989 Apr;169(2):365–376. doi: 10.1016/0042-6822(89)90162-1. [DOI] [PubMed] [Google Scholar]
- Osatomi K., Fuke I., Tsuru D., Shiba T., Sakaki Y., Sumiyoshi H. Nucleotide sequence of dengue type 3 virus genomic RNA encoding viral structural proteins. Virus Genes. 1988 Oct;2(1):99–108. doi: 10.1007/BF00569739. [DOI] [PubMed] [Google Scholar]
- Pincus S., Mason P. W., Konishi E., Fonseca B. A., Shope R. E., Rice C. M., Paoletti E. Recombinant vaccinia virus producing the prM and E proteins of yellow fever virus protects mice from lethal yellow fever encephalitis. Virology. 1992 Mar;187(1):290–297. doi: 10.1016/0042-6822(92)90317-i. [DOI] [PubMed] [Google Scholar]
- Preugschat F., Yao C. W., Strauss J. H. In vitro processing of dengue virus type 2 nonstructural proteins NS2A, NS2B, and NS3. J Virol. 1990 Sep;64(9):4364–4374. doi: 10.1128/jvi.64.9.4364-4374.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapoport T. A. Transport of proteins across the endoplasmic reticulum membrane. Science. 1992 Nov 6;258(5084):931–936. doi: 10.1126/science.1332192. [DOI] [PubMed] [Google Scholar]
- Rice C. M., Grakoui A., Galler R., Chambers T. J. Transcription of infectious yellow fever RNA from full-length cDNA templates produced by in vitro ligation. New Biol. 1989 Dec;1(3):285–296. [PubMed] [Google Scholar]
- Rice C. M., Lenches E. M., Eddy S. R., Shin S. J., Sheets R. L., Strauss J. H. Nucleotide sequence of yellow fever virus: implications for flavivirus gene expression and evolution. Science. 1985 Aug 23;229(4715):726–733. doi: 10.1126/science.4023707. [DOI] [PubMed] [Google Scholar]
- Rice C. M., Levis R., Strauss J. H., Huang H. V. Production of infectious RNA transcripts from Sindbis virus cDNA clones: mapping of lethal mutations, rescue of a temperature-sensitive marker, and in vitro mutagenesis to generate defined mutants. J Virol. 1987 Dec;61(12):3809–3819. doi: 10.1128/jvi.61.12.3809-3819.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Linares A., Cahour A., Després P., Girard M., Bouloy M. Processing of yellow fever virus polyprotein: role of cellular proteases in maturation of the structural proteins. J Virol. 1989 Oct;63(10):4199–4209. doi: 10.1128/jvi.63.10.4199-4209.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T., Takamura C., Yasuda A., Miyamoto M., Kamogawa K., Yasui K. High-level expression of the Japanese encephalitis virus E protein by recombinant vaccinia virus and enhancement of its extracellular release by the NS3 gene product. Virology. 1993 Feb;192(2):483–490. doi: 10.1006/viro.1993.1064. [DOI] [PubMed] [Google Scholar]
- Schägger H., von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987 Nov 1;166(2):368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- Shiu S. Y., Ayres M. D., Gould E. A. Genomic sequence of the structural proteins of louping ill virus: comparative analysis with tick-borne encephalitis virus. Virology. 1991 Jan;180(1):411–415. doi: 10.1016/0042-6822(91)90048-g. [DOI] [PubMed] [Google Scholar]
- Speight G., Westaway E. G. Carboxy-terminal analysis of nine proteins specified by the flavivirus Kunjin: evidence that only the intracellular core protein is truncated. J Gen Virol. 1989 Aug;70(Pt 8):2209–2214. doi: 10.1099/0022-1317-70-8-2209. [DOI] [PubMed] [Google Scholar]
- Sumiyoshi H., Mori C., Fuke I., Morita K., Kuhara S., Kondou J., Kikuchi Y., Nagamatu H., Igarashi A. Complete nucleotide sequence of the Japanese encephalitis virus genome RNA. Virology. 1987 Dec;161(2):497–510. doi: 10.1016/0042-6822(87)90144-9. [DOI] [PubMed] [Google Scholar]
- Trent D. W., Kinney R. M., Johnson B. J., Vorndam A. V., Grant J. A., Deubel V., Rice C. M., Hahn C. Partial nucleotide sequence of St. Louis encephalitis virus RNA: structural proteins, NS1, ns2a, and ns2b. Virology. 1987 Feb;156(2):293–304. doi: 10.1016/0042-6822(87)90409-0. [DOI] [PubMed] [Google Scholar]
- Wengler G., Czaya G., Färber P. M., Hegemann J. H. In vitro synthesis of West Nile virus proteins indicates that the amino-terminal segment of the NS3 protein contains the active centre of the protease which cleaves the viral polyprotein after multiple basic amino acids. J Gen Virol. 1991 Apr;72(Pt 4):851–858. doi: 10.1099/0022-1317-72-4-851. [DOI] [PubMed] [Google Scholar]
- Wengler G., Wengler G., Nowak T., Castle E. Description of a procedure which allows isolation of viral nonstructural proteins from BHK vertebrate cells infected with the West Nile flavivirus in a state which allows their direct chemical characterization. Virology. 1990 Aug;177(2):795–801. doi: 10.1016/0042-6822(90)90552-3. [DOI] [PubMed] [Google Scholar]
- Westaway E. G. Flavivirus replication strategy. Adv Virus Res. 1987;33:45–90. doi: 10.1016/s0065-3527(08)60316-4. [DOI] [PubMed] [Google Scholar]
- Yamshchikov V. F., Compans R. W. Regulation of the late events in flavivirus protein processing and maturation. Virology. 1993 Jan;192(1):38–51. doi: 10.1006/viro.1993.1006. [DOI] [PubMed] [Google Scholar]
- Yasuda A., Kimura-Kuroda J., Ogimoto M., Miyamoto M., Sata T., Sato T., Takamura C., Kurata T., Kojima A., Yasui K. Induction of protective immunity in animals vaccinated with recombinant vaccinia viruses that express PreM and E glycoproteins of Japanese encephalitis virus. J Virol. 1990 Jun;64(6):2788–2795. doi: 10.1128/jvi.64.6.2788-2795.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Mohan P. M., Padmanabhan R. Processing and localization of Dengue virus type 2 polyprotein precursor NS3-NS4A-NS4B-NS5. J Virol. 1992 Dec;66(12):7549–7554. doi: 10.1128/jvi.66.12.7549-7554.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B. T., Prince G., Horswood R., Eckels K., Summers P., Chanock R., Lai C. J. Expression of dengue virus structural proteins and nonstructural protein NS1 by a recombinant vaccinia virus. J Virol. 1987 Dec;61(12):4019–4022. doi: 10.1128/jvi.61.12.4019-4022.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Mackow E., Buckler-White A., Markoff L., Chanock R. M., Lai C. J., Makino Y. Cloning full-length dengue type 4 viral DNA sequences: analysis of genes coding for structural proteins. Virology. 1986 Nov;155(1):77–88. doi: 10.1016/0042-6822(86)90169-8. [DOI] [PubMed] [Google Scholar]