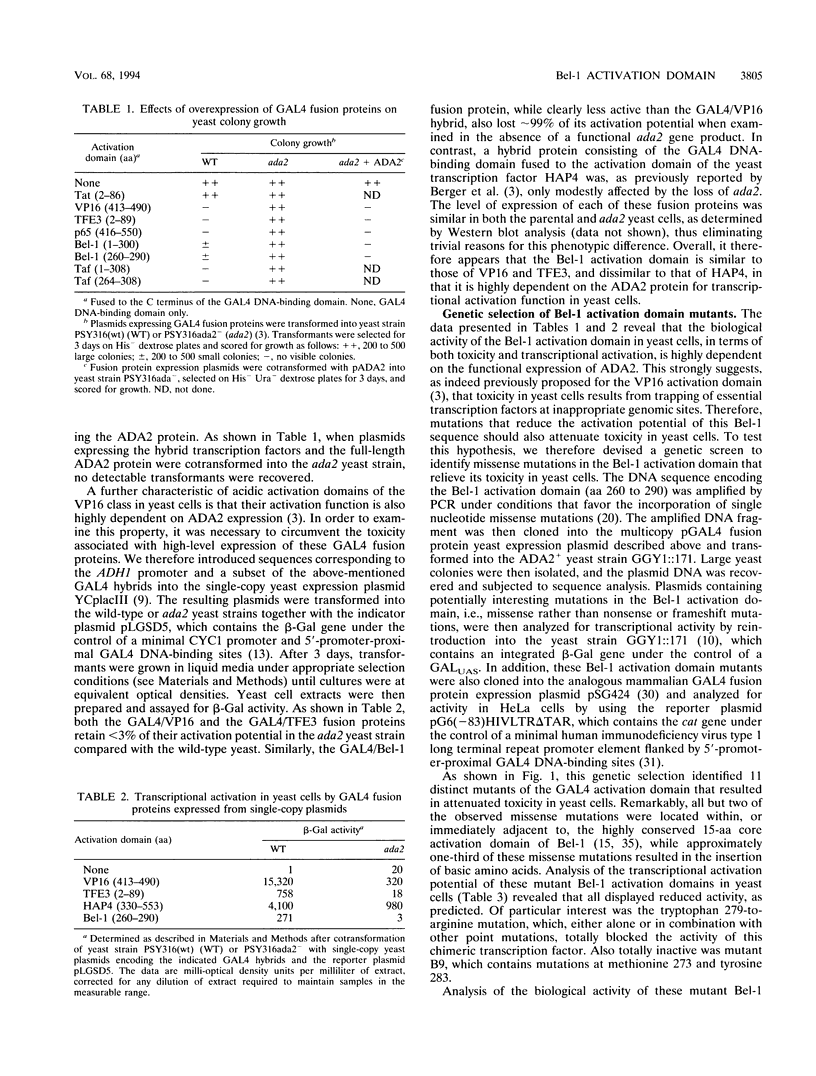
Abstract
Human foamy virus encodes a nuclear regulatory protein, termed Bel-1, that serves as a potent activator of viral transcription. Mutational analysis has identified a small, discrete activation domain within Bel-1 that is highly active in both higher and lower eukaryotic cells. Here, we demonstrate that the activation domain of Bel-1 is highly dependent on the ADA2 transcriptional adaptor for biological activity in yeast cells, a property previously shown to be a hallmark of the VP16 class of acidic transcriptional activators (S. L. Berger, B. Pina, N. Silverman, G. A. Marcus, J. Agapite, J. L. Regier, S. J. Triezenberg, and L. Guarente, Cell 70:251-265, 1992). Using genetic selection in yeast cells, we have derived a set of point mutants within the Bel-1 activation domain that display a qualitatively similar loss in activation potential when examined in either yeast or human cells. These data indicate that the Bel-1 activation domain functions similarly in both lower and higher eukaryotes and strongly suggest that Bel-1 belongs to the VP16 class of acidic transcription factors.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ballard D. W., Dixon E. P., Peffer N. J., Bogerd H., Doerre S., Stein B., Greene W. C. The 65-kDa subunit of human NF-kappa B functions as a potent transcriptional activator and a target for v-Rel-mediated repression. Proc Natl Acad Sci U S A. 1992 Mar 1;89(5):1875–1879. doi: 10.1073/pnas.89.5.1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann H., Su L. K., Kadesch T. TFE3: a helix-loop-helix protein that activates transcription through the immunoglobulin enhancer muE3 motif. Genes Dev. 1990 Feb;4(2):167–179. doi: 10.1101/gad.4.2.167. [DOI] [PubMed] [Google Scholar]
- Berger S. L., Piña B., Silverman N., Marcus G. A., Agapite J., Regier J. L., Triezenberg S. J., Guarente L. Genetic isolation of ADA2: a potential transcriptional adaptor required for function of certain acidic activation domains. Cell. 1992 Jul 24;70(2):251–265. doi: 10.1016/0092-8674(92)90100-q. [DOI] [PubMed] [Google Scholar]
- Bogerd H. P., Fridell R. A., Blair W. S., Cullen B. R. Genetic evidence that the Tat proteins of human immunodeficiency virus types 1 and 2 can multimerize in the eukaryotic cell nucleus. J Virol. 1993 Aug;67(8):5030–5034. doi: 10.1128/jvi.67.8.5030-5034.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy B., Green M. R. Eukaryotic activators function during multiple steps of preinitiation complex assembly. Nature. 1993 Dec 9;366(6455):531–536. doi: 10.1038/366531a0. [DOI] [PubMed] [Google Scholar]
- Cress W. D., Triezenberg S. J. Critical structural elements of the VP16 transcriptional activation domain. Science. 1991 Jan 4;251(4989):87–90. doi: 10.1126/science.1846049. [DOI] [PubMed] [Google Scholar]
- Cullen B. R. Use of eukaryotic expression technology in the functional analysis of cloned genes. Methods Enzymol. 1987;152:684–704. doi: 10.1016/0076-6879(87)52074-2. [DOI] [PubMed] [Google Scholar]
- Garrett E. D., He F., Bogerd H. P., Cullen B. R. Transcriptional trans activators of human and simian foamy viruses contain a small, highly conserved activation domain. J Virol. 1993 Nov;67(11):6824–6827. doi: 10.1128/jvi.67.11.6824-6827.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz R. D., Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988 Dec 30;74(2):527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Gill G., Ptashne M. Mutants of GAL4 protein altered in an activation function. Cell. 1987 Oct 9;51(1):121–126. doi: 10.1016/0092-8674(87)90016-x. [DOI] [PubMed] [Google Scholar]
- Goding C. R., O'Hare P. Herpes simplex virus Vmw65-octamer binding protein interaction: a paradigm for combinatorial control of transcription. Virology. 1989 Dec;173(2):363–367. doi: 10.1016/0042-6822(89)90548-5. [DOI] [PubMed] [Google Scholar]
- Goodrich J. A., Hoey T., Thut C. J., Admon A., Tjian R. Drosophila TAFII40 interacts with both a VP16 activation domain and the basal transcription factor TFIIB. Cell. 1993 Nov 5;75(3):519–530. doi: 10.1016/0092-8674(93)90386-5. [DOI] [PubMed] [Google Scholar]
- Guarente L., Yocum R. R., Gifford P. A GAL10-CYC1 hybrid yeast promoter identifies the GAL4 regulatory region as an upstream site. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7410–7414. doi: 10.1073/pnas.79.23.7410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn S. Structure(?) and function of acidic transcription activators. Cell. 1993 Feb 26;72(4):481–483. doi: 10.1016/0092-8674(93)90064-w. [DOI] [PubMed] [Google Scholar]
- He F., Sun J. D., Garrett E. D., Cullen B. R. Functional organization of the Bel-1 trans activator of human foamy virus. J Virol. 1993 Apr;67(4):1896–1904. doi: 10.1128/jvi.67.4.1896-1904.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keegan L., Gill G., Ptashne M. Separation of DNA binding from the transcription-activating function of a eukaryotic regulatory protein. Science. 1986 Feb 14;231(4739):699–704. doi: 10.1126/science.3080805. [DOI] [PubMed] [Google Scholar]
- Keller A., Partin K. M., Löchelt M., Bannert H., Flügel R. M., Cullen B. R. Characterization of the transcriptional trans activator of human foamy retrovirus. J Virol. 1991 May;65(5):2589–2594. doi: 10.1128/jvi.65.5.2589-2594.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lee K. J., Lee A. H., Sung Y. C. Multiple positive and negative cis-acting elements that mediate transactivation by bel1 in the long terminal repeat of human foamy virus. J Virol. 1993 Apr;67(4):2317–2326. doi: 10.1128/jvi.67.4.2317-2326.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuther K. K., Salmeron J. M., Johnston S. A. Genetic evidence that an activation domain of GAL4 does not require acidity and may form a beta sheet. Cell. 1993 Feb 26;72(4):575–585. doi: 10.1016/0092-8674(93)90076-3. [DOI] [PubMed] [Google Scholar]
- Löchelt M., Zentgraf H., Flügel R. M. Construction of an infectious DNA clone of the full-length human spumaretrovirus genome and mutagenesis of the bel 1 gene. Virology. 1991 Sep;184(1):43–54. doi: 10.1016/0042-6822(91)90820-2. [DOI] [PubMed] [Google Scholar]
- Mergia A., Renshaw-Gegg L. W., Stout M. W., Renne R., Herchenröeder O. Functional domains of the simian foamy virus type 1 transcriptional transactivator (Taf). J Virol. 1993 Aug;67(8):4598–4604. doi: 10.1128/jvi.67.8.4598-4604.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mergia A., Shaw K. E., Pratt-Lowe E., Barry P. A., Luciw P. A. Identification of the simian foamy virus transcriptional transactivator gene (taf). J Virol. 1991 Jun;65(6):2903–2909. doi: 10.1128/jvi.65.6.2903-2909.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regier J. L., Shen F., Triezenberg S. J. Pattern of aromatic and hydrophobic amino acids critical for one of two subdomains of the VP16 transcriptional activator. Proc Natl Acad Sci U S A. 1993 Feb 1;90(3):883–887. doi: 10.1073/pnas.90.3.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder R. G. The complexities of eukaryotic transcription initiation: regulation of preinitiation complex assembly. Trends Biochem Sci. 1991 Nov;16(11):402–408. doi: 10.1016/0968-0004(91)90164-q. [DOI] [PubMed] [Google Scholar]
- Sadowski I., Ptashne M. A vector for expressing GAL4(1-147) fusions in mammalian cells. Nucleic Acids Res. 1989 Sep 25;17(18):7539–7539. doi: 10.1093/nar/17.18.7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southgate C. D., Green M. R. The HIV-1 Tat protein activates transcription from an upstream DNA-binding site: implications for Tat function. Genes Dev. 1991 Dec;5(12B):2496–2507. doi: 10.1101/gad.5.12b.2496. [DOI] [PubMed] [Google Scholar]
- Stringer K. F., Ingles C. J., Greenblatt J. Direct and selective binding of an acidic transcriptional activation domain to the TATA-box factor TFIID. Nature. 1990 Jun 28;345(6278):783–786. doi: 10.1038/345783a0. [DOI] [PubMed] [Google Scholar]
- Venkatesh L. K., Chinnadurai G. The carboxy-terminal transcription enhancement region of the human spumaretrovirus transactivator contains discrete determinants of the activator function. J Virol. 1993 Jul;67(7):3868–3876. doi: 10.1128/jvi.67.7.3868-3876.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh L. K., Theodorakis P. A., Chinnadurai G. Distinct cis-acting regions in U3 regulate trans-activation of the human spumaretrovirus long terminal repeat by the viral bel1 gene product. Nucleic Acids Res. 1991 Jul 11;19(13):3661–3666. doi: 10.1093/nar/19.13.3661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernet T., Dignard D., Thomas D. Y. A family of yeast expression vectors containing the phage f1 intergenic region. Gene. 1987;52(2-3):225–233. doi: 10.1016/0378-1119(87)90049-7. [DOI] [PubMed] [Google Scholar]




