Abstract
Response to the steroid hormone ecdysone in Drosophila is controlled by genetic regulatory hierarchies that include eight members of the nuclear receptor protein family. The DHR3 gene, located within the 46F early-late ecdysone-inducible chromosome puff, encodes an orphan nuclear receptor that recently has been shown to exert both positive and negative regulatory effects in the ecdysone-induced genetic hierarchies at metamorphosis. We used a reverse genetics approach to identify 11 DHR3 mutants from a pool of lethal mutations in the 46F region on the second chromosome. Two DHR3 mutations result in amino acid substitutions within the conserved DNA binding domain. Analysis of DHR3 mutants reveals that DHR3 function is required to complete embryogenesis. All DHR3 alleles examined result in nervous system defects in the embryo.
Pulses of the steroid hormone 20-hydroxyecdysone (referred to here as ecdysone) occur throughout the Drosophila life cycle and act to coordinate and regulate multiple developmental transitions (1). The most dramatic of these transitions is the larval to adult metamorphosis where most, if not all, tissues of the larva respond to ecdysone. Ecdysone acts through a heteromeric ecdysone receptor consisting of two nuclear receptors, EcR and USP (2–5). The genetic regulatory networks that operate downstream of the ecdysone receptor are best understood for the larval salivary gland during metamorphosis by virtue of the ecdysone-induced transcription puffs visible in this tissue. In vitro studies of the induction and regression of salivary gland transcription puffs have led to a regulatory model in which a small set of early puffs, induced directly and immediately by ecdysone, produce a product (or products) P that acts both to induce a larger set of late puffs and to repress early puffs (6). A small subset of the late puffs (the early-late puffs) respond similarly to the early puffs in ecdysone addition and withdrawal experiments (7). Subsequent molecular analysis has shown that early-late class genes, like the early genes, are directly induced by ecdysone, although they require protein synthesis for their maximal induction (8, 9).
Recent studies of the DHR3 early-late gene, which encodes an orphan nuclear receptor (10), suggest that it plays a key role in the ecdysone regulatory hierarchies at metamorphosis (11, 12). Overexpression of DHR3 in vivo and in vitro results in repression of the early genes E74A, E75A, and BR-C and premature induction of βFTZ-F1, a gene that regulates prepupal ecdysone responsive genes (13). These results have led to the proposal that DHR3, like P, has a dual regulatory function, acting negatively early in ecdysone response to repress early gene expression and later acting positively to activate the βFTZ-F1 gene (11, 12). Biochemical evidence suggests possible mechanisms for DHR3 regulatory effects. DHR3 may repress early genes in a DNA binding domain independent fashion through contact with EcR, whereas the timing of βFTZ-F1 activation may be determined by the physical interaction of DHR3 and E75B, a nuclear receptor lacking a complete DNA binding domain (12).
The proposed functions for DHR3 lead to the testable predictions that inactivation of DHR3 at the proper time during metamorphosis will lead to failures of early gene repression and βFTZ-F1 induction. A prerequisite for testing these predictions is the isolation of DHR3 mutants. Here we report identification of DHR3 loss-of-function mutations. We have used denaturing gradient gel electrophoresis (DGGE) to identify a complementation group consisting of 11 DHR3 alleles from among a collection of lethal mutations mapping to the 46F chromosomal region. Two DHR3 mutations are missense mutations within the DNA binding domain. All DHR3 mutants tested are embryonic lethal and have nervous system defects. The demonstration of an embryonic function for DHR3 is consistent with the expression of DHR3 during embryogenesis (10) and, together with similar temporal requirements for EcR, usp, and E75 (M.B., F. B. Imam, W. S. Talbot, B. Ganetzky, and D. S. Hogness, unpublished results, and refs. 14–16), suggests important embryonic functions for ecdysone.
MATERIALS AND METHODS
DGGE.
Genomic DNA was isolated from flies heterozygous for 46E-F lethal mutants or the adh cn pr parental chromosome and the CyO balancer chromosome. A 234-nt DNA fragment encoding the DHR3 DNA binding domain was amplified by PCR using primers specific to this region. The 5′ primer (DH1) has the sequence 5′-CG-GCTCAAATTGAGATAATTCC-3′, where CG- represents a 40-nt GC clamp designed to prevent complete denaturation of products during electrophoresis (17). The 3′ primer (DH2) has the sequence 5′-ACCATCACGGCTCATTCC-3′. PCR products were electrophoresed at 60°C on 6.5% acrylamide gels containing an increasing gradient of denaturant (18) and were visualized by ethidium bromide staining.
The 60 46E-F lethals were previously isolated and placed into complementation groups (19). Of these 60 candidate lethals, 47 were screened by DGGE. Two poorly growing members of the large A complementation group and the 11 members of the B complementation group, which corresponds to the Drosophila phosphofructokinase gene (20), were not screened.
Sequence Analysis.
DNA from DHR3R107G and DHR3G60S strains was PCR-amplified, and DGGE was performed as described. In each case, the most rapidly migrating band (see Fig. 2) represents the mutant homoduplex species. These bands were excised from the gels and reamplified using DH1 and DH2 primers. DNA from adh cn pr was PCR-amplified as above and sequenced. The entire procedure was performed two independent times to control for possible PCR amplification errors. Sequencing of both strands of each template was performed in the Molecular Genetics Instrumentation Facility at the University of Georgia on an Applied Biosystems 373 stretch machine using the manufacturer’s standard protocol for dye terminator sequencing.
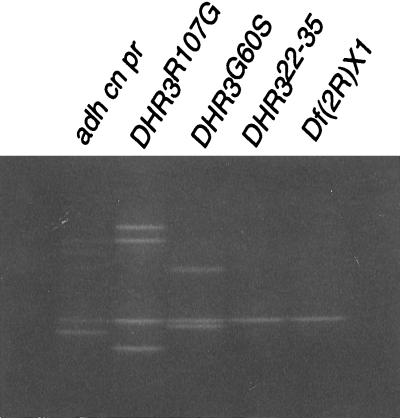
Figure 2.
Identification of DHR3 mutations by DGGE. The genomic region encoding the DHR3 DNA binding domain was PCR-amplified, separated on acrylamide denaturing gradient gels, and stained with ethidium bromide. All samples are from strains heterozygous for the balancer chromosome CyO. PCR products from the D complementation group lethals DHR3R107G and DHR3G60S have band shifts relative to the parental chromosome adh cn pr. The PCR product from lethal DHR322-35 has a pattern identical to the DHR3 deficiency Df(2R)X1.
Lethal Phase Analysis.
DHR3 mutant animals were marked with yellow (y) and maintained as heterozygotes with the second chromosome balancer CyO,y+ (a gift of J. Botas, Baylor, Houston). Twenty five yw;DHR3/Cyo,y+ males were mated with 25 yw;Df(2R)12/CyO,y+ (see Fig. 1) virgin females at 25°C for 2 to 4 days before egg collections were begun. After a 6-hr egg collection on grape juice agar plates, 200 eggs from each cross were isolated. Thirty-six hours after the end of the egg collection period, the plates were examined to determine the number of hatched eggs and living y first instar larvae. As necessary, living y larvae were followed through all stages of development. Two DHR3 mutant chromosomes, DHR325-74 and DHR366-20, did not survive over CyO,y+ and were not tested.
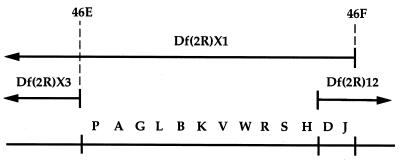
Figure 1.
Lethal mutations in the 46EF region. Thirteen lethal complementation groups (designated by a letter between A and W) that are included within Df(2R)X1 but excluded by Df(2R)X3 are shown at the bottom. Two of these groups (D and J) also are included in Df(2R)12, a small deficiency generated by x-ray mutagenesis of a P-element insert at 47A (R. Burgess and T. Schwarz, personal communication and ref. 39), and therefore are placed at the right end of this interval. The cytological endpoints of Df(2R)X3 and Df(2R)X1 are shown at the top.
Immunostaining of Embryos.
Males homozygous for adh cn pr or heterozygous for either DHR3G60S, DHR322-35, DHR391-9, DHR322-12, DHR312-6, DHR330-45, or DHR3R107G and the CyO,wg-lacZ balancer (a gift of D. Bilder, Stanford) were mated to Df(2R)X1/CyO,wg-lacZ virgin females. We did not test the remaining four DHR3 alleles because of their inability to be maintained as heterozygotes with the CyO,wg-lacZ balancer. In addition to 10 DHR3/Df(2R)X1 embryos of each genotype, we examined wild-type siblings from each cross to control for dominant effects from the deficiency or balancer chromosomes. We examined CyO,wg-lacZ in a Canton S background as a second control for dominant effects from this chromosome.
Eggs were collected for 5-hr intervals and allowed to grow at 25°C until 11–16 hr after egg laying. Embryos were dechorionated, fixed in heptane/formaldehyde, and devitellinized by vigorous mixing in heptane/methanol. The embryos subsequently were rehydrated through several washes in PBT (PBS + 0.5% Triton X-100) and then incubated in blocking solution (5% goat serum) for 2 hr. Incubation with mouse mAbs 22C10 (21, 22) or BP102 (23) (1:5 dilution) was performed in 5% goat serum overnight at 4°C in the presence of mAb for β-galactosidase detection (Promega). Only homozygous DHR3 mutants will lack β-galactosidase staining.
Embryos were rinsed four times with PBT and incubated for 2 hr at room temperature with a 150-fold dilution of goat α-mouse IgG horseradish peroxidase (HRP) conjugated secondary antibody (Promega). HRP detection was performed with diaminobenzidine in the presence of 8% NiCl2. Animals were viewed by differential interference contrast microscopy optics using a Zeiss axiophot microscope.
For DHR3G60S, DHR3R107G, and DHR322-35 a similar protocol was followed for a Drosophila muscle myosin (24) antibody. Secondary detection was performed with tetramethylrhodamine B isothiocyanate- and fluorescein isothiocyanate-conjugated antibodies.
RESULTS
Identification of DHR3 Mutants.
The DHR3 gene has been mapped to the 46F position on chromosome 2R by in situ hybridization (10). A chromosomal deficiency that removes the 46C-F region, Df(2R)X1 (25), was used to isolate 126 lethal mutants induced with ethylmethane-sulfonate, diepoxybutane, or gamma irradiation (19). Sixty of these mutations fall into 13 complementation groups in the 46E-F region (Fig. 1) because they complement Df(2R)X3, a deficiency for the 46C-E region (25).
To determine which complementation group corresponds to DHR3, we performed denaturing gradient gel electrophoresis, a rapid and sensitive technique capable of detecting point mutations and small deletions (18), to screen the 46E-F lethal mutations. We chose to screen the DHR3 DNA binding domain for mutations because this domain is the most strongly conserved region in the nuclear receptor family (26). Primers specific to sequences encoding the DHR3 DNA binding domain (see Materials and Methods) were used to amplify genomic DNA from animals heterozygous for a 46E-F lethal and the CyO balancer chromosome. PCR products were analyzed on denaturing gradient gels for changes in mobility relative to the adh cn pr parental chromosome.
Two alleles from complementation group D, DHR3R107G and DHR3G60S (Fig. 2), have bands with altered electrophoretic mobility. In addition, a third member of the D group, DHR322-35, has a DGGE pattern identical to that of Df(2R)X1 (Fig. 2), a deletion that encompasses DHR3. This observation is consistent with a DNA lesion on the DHR322-35 chromosome that deletes or separates the PCR priming sites. The remaining 46E-F lethal mutations exhibit a pattern identical to that of the parental chromosome (data not shown). These results identify the D group, consisting of 11 alleles, as the DHR3 complementation group.
Two DHR3 Mutations Affect Amino Acid Residues in the DNA Binding Domain.
The most striking feature of the DNA binding domain of nuclear receptor proteins is two highly conserved cysteine-rich regions that function as zinc coordination modules. These modules supply target DNA contact sites as well as dimerization interfaces with other family members (27). Because the observed electrophoretic patterns for DHR3R107G and DHR3G60S (Fig. 2) were suggestive of point mutations or small deletions within the exon encoding the DNA binding domain, we sequenced this region from these alleles. The DNA sequence changes and predicted amino acid changes of these alleles are shown in Fig. 3A.
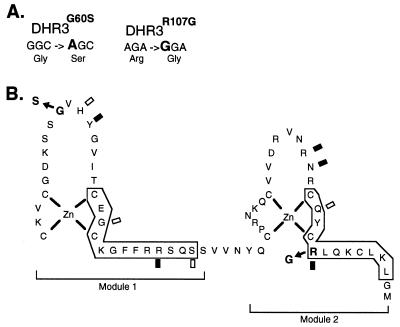
Figure 3.
DHR3 mutations within the DNA binding domain. (A) The DNA sequence and predicted amino acid changes for DHR3G60S and DHR3R107G are shown. (B) A predicted Zn coordination scheme for the DHR3 DNA binding domain based upon known nuclear receptor DNA binding domain crystal structures (28–30) is shown as are the amino acid changes for DHR3G60S (G to S change in the first zinc module) and DHR3R107G (R to G change in the second zinc module). Amino acid numbering is as in ref. 10. Specific phosphate backbone contact sites common to the four known DNA binding domain crystal structures are indicated by shaded rectangles. Specific phosphate backbone contacts conserved in two or three structures are indicated by open rectangles. α-helical regions are boxed.
The DHR3G60S allele has a single change at nucleotide 404 (numbering of nucleotides is according to the cDNA sequence presented in ref. 10) predicted to substitute a serine for a glycine residue at the tip of module 1 (Fig. 3B). This substitution occurs adjacent to amino acids shown to mediate phosphate backbone contacts in structural studies of other nuclear receptor DNA binding domains (Fig. 3B and refs. 28–30). The serine for glycine substitution may affect the DHR3 protein fold and thus interfere with these contacts. The glycine residue at this position is strongly conserved in the nuclear receptor superfamily (31), and a missense mutation that affects this residue in the vitamin D receptor has been reported in a patient with rickets syndrome (32). The same residue of the androgen receptor is altered in a patient with androgen insensitivity syndrome (32).
The change at nucleotide 545 in DHR3R107G is predicted to cause substitution of glycine for an arginine in module 2 (Fig. 3B). The arginine at this position is universally conserved in the nuclear receptor superfamily (31) and has been shown to contact the phosphate backbone in studies of other nuclear receptor protein structures (Fig. 3B and refs. 28–30). Missense mutations that affect the arginine residue at this position in the vitamin D and androgen receptors have been reported for rickets and androgen insensitivity patients, respectively (32).
DHR3 Function Is Required During Embryogenesis.
To determine the phenotype of the DHR3 mutants, we first defined the time of death of each mutant when heterozygous for Df(2R)12, a deletion that fails to complement all DHR3 alleles. The percent survival of each mutant relative to the adh cn pr chromosome is shown in Table 1. The predominant lethal phase of all DHR3 mutants tested is embryogenesis (Table 1), with very few (six strains) or no (three strains) first instar larvae detected. For a single strain (DHR391-9), one animal survived to the third larval instar. To confirm these results, we scored larger egg collections (>500 eggs) in an independent set of experiments for three DHR3 mutants (DHR312-6, DHR3G60S, and DHR391-9). In these experiments, one (DHR312-6 and DHR391-9) or two (DHR3G60S) hatched y first instar larvae were detected and all y larvae died during the first larval instar. Thus, our data indicate that DHR3 function is required during embryogenesis.
Table 1.
DHR3 lethal phase
| Paternal allele | Percent survival
|
|||||
|---|---|---|---|---|---|---|
| Hatched | 1st instar | 2nd instar | 3rd instar | Prepupae | Adult | |
| adh cn pr | 100 | 100 | 100 | 97 | 97 | 97 |
| DHR3G60S | 0 | 0 | 0 | 0 | 0 | 0 |
| DHR3R107G | 3 | 0 | 0 | 0 | 0 | 0 |
| DHR322-35 | 0 | 0 | 0 | 0 | 0 | 0 |
| DHR312-6 | 6 | 0 | 0 | 0 | 0 | 0 |
| DHR314-4 | 9 | 0 | 0 | 0 | 0 | 0 |
| DHR322-12 | 12 | 0 | 0 | 0 | 0 | 0 |
| DHR330-45 | 0 | 0 | 0 | 0 | 0 | 0 |
| DHR351-10 | 3 | 0 | 0 | 0 | 0 | 0 |
| DHR391-9 | 3 | 3 | 3 | 3 | 0 | 0 |
DHR3 mutants heterozygous for Df(2R)12 were scored at six times during development (see Materials and Methods). Percent survival is expressed as a percentage of the y animals produced in the adh cn pr control cross (33 animals) from a collection of 200 eggs at the following stages: hatching; first, second, and third larval; prepupal; and adult.
DHR3 Mutants Have Nervous System Defects.
To understand the nature of the observed embryonic lethality, we first examined the external structure of DHR3 embryos. Cuticle preparations from 24- to 36-hr embryos indicate no obvious morphological differences between wild-type and DHR3G60S, DHR3R107G, or DHR322-35 embryos that are heterozygous for either Df(2R)X1 or Df(2R)12 (data not shown). Thus, DHR3 has no apparent function in patterning or cuticle deposition.
Lam et al. (11) have shown that DHR3 is widely expressed during embryogenesis in tissues that include the gut, salivary gland, epidermis, and, at low levels, the ventral nerve cord. Because DHR3 is expressed widely, we chose antibody markers that would allow us to examine a variety of internal structures of the embryo. We used antibodies directed against the nervous system, mAbs 22C10 (21, 22) and BP102 (23), or directed against Drosophila muscle myosin (24) and screened for detectable structural changes in DHR3/Df(2R)X1 embryos (see Materials and Methods). DHR3/Df(2R)X1 embryos were identified by lack of staining with an antibody directed against the β-galactosidase protein expressed from the CyO,wg-lacZ balancer chromosome.
Peripheral neurogenesis in Drosophila is complete by 14 hr of development (33). At this time, the peripheral nervous system (PNS) has a stereotypical and well defined pattern (33, 34), which can be observed by staining with mAb 22C10. This antibody stains 44 neurons and their projections in each abdominal hemisegment as well as a similar number in each of the three thoracic hemisegments. Within each hemisegment the neurons are arranged within invariant clusters designated as dorsal, lateral, or ventral clusters (Fig. 4D).
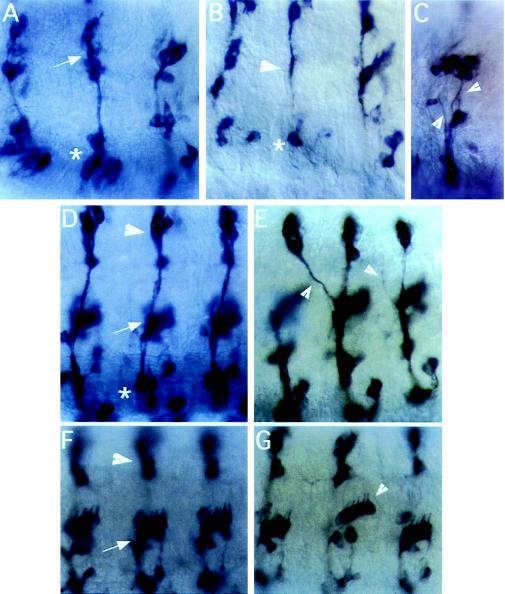
Figure 4.
PNS phenotypes of DHR3 mutant embryos. (A–G) Lateral views of 11- to 16-hr embryos stained with mAb 22C10. Anterior is to the left, dorsal to the top. (A, D, and F) Wild-type animals. (B, C, E, and G) DHR3/Df(2R)X1 animals. (A) Lateral (arrow) and ventral clusters (asterisk) of segments T2, T3, and A1. (B) DHR3R107G/Df(2R)X1 animal lacking neurons in the lateral (arrowhead) and ventral (asterisk) clusters. (C) Lateral and ventral clusters of the A2 segment of a DHR391-9/Df(2R)X1 embryo, which has severe fasciculation defects (arrowheads). (D) Dorsal (arrowhead), lateral (arrow), and ventral clusters (asterisk) for A2-A4. (E) A similar view of a DHR312-6/Df(2R)X1 embryo with defects in axonal projections (arrowheads). (F and G) Views of dorsal (arrowhead in F) and lateral (arrow in F) abdominal clusters. (G) One lateral cluster of chordotonal neurons (indicated by an arrowhead) in this DHR391-9/Df(2R)X1 animal is shifted dorsally relative to similar clusters in neighboring segments.
We tested DHR3 mutants for alterations in PNS patterning by immunostaining with mAb 22C10. For each genotype examined, approximately 50% of the embryos contained PNS defects. The types of defects noted included loss of neurons, pathfinding and fasciculation defects, cluster organization defects, and displacement of neurons to inappropriate positions within the PNS. These defects were not limited to a specific cluster but were found in many thoracic and abdominal clusters.
A severely affected embryo is shown in Fig. 4B. Lateral cluster neurons are absent (Fig. 4B, arrowhead) as well as most ventral cluster neurons (Fig. 4B, asterisk). Fasciculation defects, or defects in axon bundling, were observed in a number of animals (Fig. 4C, arrowheads). In addition to the defects in fasciculation, this animal lacks organization in the lateral and ventral clusters (compare with Fig. 4 A and F). Incorrect or missing projections (Fig. 4E, arrowheads) and displacement of neurons or entire clusters of neurons (Fig. 4G, arrowhead) were observed in DHR3 animals. However, the overall architecture of the PNS usually is maintained in the mutants (Fig. 4, compare F and G).
We also examined DHR3 mutants for defects in the central nervous system (CNS). mAb BP102 specifically stains axons of CNS neurons, recognizing both commissures and longitudinal tracts (35). Examination of DHR3R107G/Df(2R)X1 embryos revealed a break in the posterior portion of the longitudinal tract in 14% of these animals (data not shown). This defect was rarely noted in other DHR3 mutant genotypes.
To examine the muscular structure of DHR3 animals we used the Drosophila α-muscle myosin antibody (24), which is specific to cardiac cells and visceral muscles as well as somatic muscle fibers (36). The pattern of muscle organization is highly sterotypical in wild-type embryos. Immunostaining revealed that DHR3/Df(2R)X1 animals have low frequency (5–15%) defects in muscle patterning that affect most or all of the somatic muscles of the body wall (data not shown). The remainder of the mutant embryos had no observable defects.
DISCUSSION
We have identified mutations in DHR3, an ecdysone-inducible early-late gene that encodes a member of the nuclear receptor superfamily. Denaturing gradient gel electrophoresis allowed rapid screening of 47 mutant strains for alterations in the DHR3 DNA binding domain exon. Sequence analysis reveals that two DHR3 alleles, DHR3R107G and DHR3G60S, have single nucleotide substitutions predicted to alter amino acids highly conserved among receptor family members. A third allele, DHR322–35, has an altered DGGE pattern that is consistent with a deletion, inversion, or translocation involving the DHR3 DNA binding domain exon. These results indicate that an 11-member complementation group in the 46E-F region corresponds to DHR3. The subsequent finding that DHR3 complementation group members can be rescued to late stages of development by transgenic expression of a DHR3 cDNA under heat-shock control (G. Lam and C. Thummel, personal communication) supports this conclusion. Other members of the DHR3 complementation group may have mutations that lie elsewhere in the gene because we did not detect an altered DGGE pattern in analysis of the DNA binding domain exon. Based on the sequence changes of the two DHR3 alleles for which the molecular defect is known and the consistent lethal period of the nine DHR3 alleles tested, we presume that most DHR3 mutants examined here have little or no DHR3 function.
Lethal phase analysis indicates that DHR3 is required to complete embryogenesis. DHR3 mutants form a normal larval cuticle and few DHR3 mutant animals survive to hatching, suggesting a late embryonic requirement for DHR3 function. This requirement is consistent with the expression of DHR3 during mid- to late embryogenesis (10, 11). Molecular studies of ecdysone response in Drosophila have focused on the function of ecdysone during metamorphosis in later development (37). Demonstration of an embryonic requirement for the DHR3 early-late gene, along with similar requirements for other key regulatory proteins that function in ecdysone response (M.B., F. B. Imam, W. S. Talbot, B. Ganetzky, and D. S. Hogness, unpublished results, and refs. 14–16), suggest additional important functions for ecdysone signaling in early development.
DHR3 mutants display defects in formation of the PNS. Using the antibody 22C10 as a marker for peripheral neurons and their projections, we found that 11- to 16-hr embryos heterozygous for Df(2R)X1 and for all DHR3 mutant alleles examined (seven alleles) have PNS defects. This phenotype, however, is incompletely penetrant as only approximately half of the animals of a given DHR3 genotype display the PNS defects. The defects include absence or mislocalization of neurons or clusters of neurons, defects in fasciculation, incorrect or missing projections, and a general disorganization of neuronal clusters. Despite localized defects, most DHR3 mutant animals maintain the sterotypical, segmental pattern of the PNS. A minor defect in central nervous system formation was detected in one DHR3 genotype (DHR3R107G/Df(2R)X1) upon staining with mAb BP102. Fourteen percent of DHR3R107G/Df(2R)X1 mutants have a break in the posterior portion of the longitudinal tract of the ventral nervous system. Minor muscle defects were detected in a similarly low percentage of DHR3 animals.
Our observations suggest that DHR3 may function in patterning of the PNS during Drosophila embryogenesis. Although DHR3 is widely expressed at this time (11) the current precision of the expression analysis does not indicate if DHR3 is expressed in peripheral neurons. The defects observed therefore may be caused by loss of DHR3 function in peripheral neurons or to loss of DHR3 function in other cell types normally involved in neuronal specification or pathfinding. Alternatively, DHR3 may have its primary function in tissues not assayed here and the PNS defects may be indirect.
Although DHR3 is essential for viability, the incomplete penetrance and variable expressivity of the DHR3 phenotype suggest that DHR3 may be partially redundant. Recently, it has been proposed that DHR3 may have overlapping functions with the E78 gene (also known as EIP78C). E78 is a second ecdysone inducible early-late gene that, unlike DHR3, is nonessential for viability (38). Isolation of DHR3 mutants will allow analysis of DHR3,E78 double mutants to address the question of functional redundancy between these genes. Double mutant analysis using mutations that affect other members of the ecdysone regulatory hierarchy also will be informative in elucidating regulatory relationships between these genes.
Recent studies based on overexpression of DHR3 during metamorphosis suggest that DHR3 has both positive and negative regulatory functions in ecdysone regulatory pathways (11, 12). DHR3 is proposed to repress the early genes E74A, E75A, and BR-C and later to activate the βFTZ-F1 gene. The early gene repression functions of DHR3 may be mediated through contact with the EcR protein and the timing of βFTZ-F1 activation through contact with E75B (12). The proposed DHR3 functions predict that loss of DHR3 function during metamorphosis will lead to failures of early gene repression and βFTZ-F1 activation. The identification of DHR3 mutations reported here will allow testing of these predictions and assessment of the regulatory functions of DHR3 during both embryonic and metamorphic responses to ecdysone.
Acknowledgments
The 22C10 monoclonal and muscle myosin polyclonal antisera were kindly provided by S. Benzer and D. Kiehart, respectively. mAb BP102, developed by C. Goodman, was provided by the Developmental Studies Hybridoma Bank maintained by the University of Iowa under contract N01-HD-7–3263 from the National Institute of Child Health and Human Development. We thank K. Bhat for aid in nervous system analysis and S. Treadway, A. E. Stephenson, and G. Gramstead for the complementation analysis. This work was supported by grants from the National Institutes of Health (GM53681 to M.B. and RR07112 to E.S.G.), the University of Georgia Research Foundation (to M.B.), the Del E. Webb foundation (to E.S.G.), a National Research Service Award for predoctoral training in genetics (to G.E.C.), and a Howard Hughes Summer Undergraduate Fellowship (to A.A.W.).
ABBREVIATIONS
- DGGE
denaturing gradient gel electrophoresis
- PNS
peripheral nervous system
References
- 1.Riddiford L M. In: The Development of Drosophila melanogaster. Bate M, Martinez-Arias A, editors. Vol. 2. Plainview, NY: Cold Spring Harbor Lab. Press; 1993. pp. 899–940. [Google Scholar]
- 2.Koelle M R, Talbot W S, Segraves W A, Bender M T, Cherbas P, Hogness D S. Cell. 1991;67:59–77. doi: 10.1016/0092-8674(91)90572-g. [DOI] [PubMed] [Google Scholar]
- 3.Koelle M R. Ph.D. thesis. Stanford, CA: Stanford University; 1992. [Google Scholar]
- 4.Yao T P, Segraves W A, Oro A E, McKeown M, Evans R M. Cell. 1992;71:63–72. doi: 10.1016/0092-8674(92)90266-f. [DOI] [PubMed] [Google Scholar]
- 5.Yao T-P, Forman B M, Jiang Z, Cherbas L, Chen J-D, McKeown M, Cherbas P, Evans R M. Nature (London) 1993;366:476–479. doi: 10.1038/366476a0. [DOI] [PubMed] [Google Scholar]
- 6.Ashburner M, Chihara C, Meltzer P, Richards G. Cold Spring Harbor Symp Quant Biol. 1974;38:655–662. doi: 10.1101/sqb.1974.038.01.070. [DOI] [PubMed] [Google Scholar]
- 7.Ashburner M, Richards G. Dev Biol. 1976;54:241–255. doi: 10.1016/0012-1606(76)90302-x. [DOI] [PubMed] [Google Scholar]
- 8.Horner M A, Chen T, Thummel C S. Dev Biol. 1995;168:490–502. doi: 10.1006/dbio.1995.1097. [DOI] [PubMed] [Google Scholar]
- 9.Stone B L, Thummel C S. Cell. 1993;75:307–320. doi: 10.1016/0092-8674(93)80072-m. [DOI] [PubMed] [Google Scholar]
- 10.Koelle M R, Segraves W A, Hogness D S. Proc Natl Acad Sci USA. 1992;89:6167–6171. doi: 10.1073/pnas.89.13.6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lam G T, Jiang C, Thummel C S. Development (Cambridge, UK) 1997;124:1757–1769. doi: 10.1242/dev.124.9.1757. [DOI] [PubMed] [Google Scholar]
- 12.White K P, Hurban P, Watanabe T, Hogness D S. Science. 1997;276:114–117. doi: 10.1126/science.276.5309.114. [DOI] [PubMed] [Google Scholar]
- 13.Woodard C T, Baehrecke E H, Thummel C S. Cell. 1994;79:607–615. doi: 10.1016/0092-8674(94)90546-0. [DOI] [PubMed] [Google Scholar]
- 14.Bilder D, Scott M P. Genetics. 1995;141:1087–1100. doi: 10.1093/genetics/141.3.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oro A E, McKeown M, Evans R M. Development (Cambridge, UK) 1992;115:449–462. doi: 10.1242/dev.115.2.449. [DOI] [PubMed] [Google Scholar]
- 16.Segraves W A. Ph.D. thesis. Stanford, CA: Stanford University; 1988. [Google Scholar]
- 17.Sheffield V C, Cox D R, Lerman L S, Myers R M. Proc Natl Acad Sci USA. 1989;86:232–236. doi: 10.1073/pnas.86.1.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Myers R M, Maniatis T, Lerman L S. Methods Enzymol. 1987;155:501–527. doi: 10.1016/0076-6879(87)55033-9. [DOI] [PubMed] [Google Scholar]
- 19.Bour B A, O’Brien M A, Lockwood W L, Goldstein E S, Bodmer R, Taghert P H, Abmayr S M, Nguyen H T. Genes Dev. 1995;9:730–741. doi: 10.1101/gad.9.6.730. [DOI] [PubMed] [Google Scholar]
- 20.Currie P D, Sullivan D T. J Biol Chem. 1994;269:24679–24687. [PubMed] [Google Scholar]
- 21.Fujita S C, Zipursky S L, Benzer S, Ferrus A, Shotwell S W. Proc Natl Acad Sci USA. 1982;79:7929–7933. doi: 10.1073/pnas.79.24.7929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zipursky S L, Venkatesh T R, Teplow D B, Benzer S. Cell. 1984;36:15–26. doi: 10.1016/0092-8674(84)90069-2. [DOI] [PubMed] [Google Scholar]
- 23.Seeger M A, Tear G, Ferres-Marco D, Goodman C S. Neuron. 1993;10:409–426. doi: 10.1016/0896-6273(93)90330-t. [DOI] [PubMed] [Google Scholar]
- 24.Kiehart D P, Feghali R. J Cell Biol. 1986;103:1517–1525. doi: 10.1083/jcb.103.4.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O’Brien M A, Roberts M S, Taghert P H. Genetics. 1994;137:121–137. doi: 10.1093/genetics/137.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Evans R M. Science. 1988;240:889–895. doi: 10.1126/science.3283939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Freedman L P, Luisi B F, Alroy A, Towers T L. In: Steroid Hormone Receptors: Basic and Clinical Aspects. Moudgil V K, editor. Boston: Birkhauser; 1993. pp. 47–73. [Google Scholar]
- 28.Luisi B F, Xu W-X, Otwinowski Z, Freedman L P, Yamamoto K R, Sigler P B. Nature (London) 1991;352:497–505. doi: 10.1038/352497a0. [DOI] [PubMed] [Google Scholar]
- 29.Rastinejad F, Perlmann T, Evans R M, Sigler P B. Nature (London) 1995;375:203–211. doi: 10.1038/375203a0. [DOI] [PubMed] [Google Scholar]
- 30.Schwabe J W R, Chapman L, Finch J T, Rhodes D. Cell. 1993;75:567–578. doi: 10.1016/0092-8674(93)90390-c. [DOI] [PubMed] [Google Scholar]
- 31.Freedman L P. Endocrinol Rev. 1992;13:129–145. doi: 10.1210/edrv-13-2-129. [DOI] [PubMed] [Google Scholar]
- 32.Luisi B F, Schwabe J W R, Freedman L P. In: Vitamins and Hormones. Litwak G, editor. Vol. 49. New York: Academic; 1994. pp. 1–47. [DOI] [PubMed] [Google Scholar]
- 33.Bodmer R, Jan Y N. Roux Arch Dev Biol. 1987;196:69–77. doi: 10.1007/BF00402027. [DOI] [PubMed] [Google Scholar]
- 34.Ghysen A, Dambly-Chaudiere C, Aceves E, Jan L Y, Jan Y N. Roux Arch Dev Biol. 1986;195:281–289. doi: 10.1007/BF00376060. [DOI] [PubMed] [Google Scholar]
- 35.Patel N H. In: Drosophila melanogaster: Practical Uses in Cell and Molecular Biology. Goldstein L S B, Fyrberg E A, editors. San Diego: Academic; 1994. pp. 445–487. [Google Scholar]
- 36.Salzberg A, Golden K, Bodmer R, Bellen H J. Genetics. 1996;144:183–196. doi: 10.1093/genetics/144.1.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thummel C S. Trends Genet. 1996;12:306–310. doi: 10.1016/0168-9525(96)10032-9. [DOI] [PubMed] [Google Scholar]
- 38.Russell S R H, Heimbeck G, Goddard C M, Carpenter A T C, Ashburner M. Genetics. 1996;144:159–170. doi: 10.1093/genetics/144.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weber U, Siegel V, Mlodzik M. EMBO J. 1995;14:6247–6257. doi: 10.1002/j.1460-2075.1995.tb00315.x. [DOI] [PMC free article] [PubMed] [Google Scholar]