Abstract
The X and Y chromosomes of the mouse, like those of other mammals, are heteromorphic over most of their length, but at the distal ends of the chromosomes is a region of sequence identity, the pseudoautosomal region (PAR), where the chromosomes pair and recombine during male meiosis. The point at which the PAR diverges into X- and Y-specific sequences is called the pseudoautosomal boundary. We have completed a genomic walk from the X-specific Amelogenin gene to the PAR. Analysis of this region revealed that the pseudoautosomal boundary of mice is located within an intron of a transcribed gene that encodes a novel RING finger protein. The first three of the exons of the gene are located on the X chromosome whereas the 3′ exons of the gene are located on both X and Y chromosomes. This unusual arrangement may indicate that the gene is in a state of transition from pseudoautosomal to X-unique and provides evidence for a process of attrition of the pseudoautosomal region on the Y chromosome.
The X and Y chromosomes of mammals are thought to have evolved from an identical pair of autosomes but since the acquisition of the sex-determining gene, the homology of the Y chromosome with the X has been almost completely eroded (1). Only a small region of identity is retained that is responsible for pairing and recombination between the chromosomes. Genetic analysis has demonstrated that loci within this region are capable of exchange between the X and Y, unlike the nonhomologous portions of these chromosomes, and therefore they behave like small autosomes. However, these loci show varying degrees of sex linkage and are thus pseudoautosomal (2).
The pseudoautosomal boundary is the point at which the pseudoautosomal region, where the X and Y chromosomes are identical, diverges into X-specific and Y-specific sequences and is therefore the proximal limit to legitimate X and Y recombination (3). Genetic analysis in humans has revealed that the degree of sex-linkage of any pseudoautosomal locus is dependent upon its distance from the boundary: the further away the locus the weaker the linkage (4). Therefore, whilst the position of the crossover between the X and Y chromosomes is random, the exchange of loci close to the boundary is a relatively rare event (3).
The nonhomologous portion of the X chromosome undergoes X-inactivation in female cells so that the gene dosage within this region is equalized between XY males and XX females. Because both males and females have two copies of all pseudoautosomal genes there is no requirement for dosage compensation and all of the human pseudoautosomal region (PAR) genes, and the single mouse PAR gene, that have been examined so far have proved to escape X-inactivation (5–8). This raises the question of whether escape of the X-inactivating signal is a property of the genes themselves or whether there is a block situated at or near the PAR boundary that prevents the spread of the X-inactivation machinery.
The mouse PAR appears to be of distinct evolutionary origin to the human PAR. None of the genes that localize to the human PAR appear to map to the mouse PAR; on the contrary these genes, when mapped in the mouse, are located on autosomes (9–10). Furthermore the human homologue of the only mouse PAR gene cloned, Steroid Sulfatase (Sts), is located just proximal to the boundary within the X-unique portion (11). It has been suggested that this lack of similarity between the PARs of different mammalian species can be explained by the repeated addition of autosomal segments onto the PAR during the course of mammalian evolution together with continual attrition or loss of pseudoautosomal material from the Y chromosome (12). To study the structure and evolution of the mouse PAR we have identified sequences flanking the pseudoautosomal boundary. Intriguingly, we found that the boundary was located within an intron of a transcribed gene. We discuss the implications of this finding in understanding the evolution of PARs in mammals.
MATERIALS AND METHODS
STS PCR Markers and YAC Libraries.
The YAC libraries used were the Medical Research Council Human Genome Mapping Programme combined mouse YAC library from which the yeast artificial chromosome (YAC) IɛU18 was isolated and the MIT mouse YAC library (through Research Genetics, Huntsville, AL) from which all the other YACs were obtained. Superpools were screened with primer pairs using PCR conditions of 45 cycles of 94°C for 30s, 52°C for 30s and 72°C for 60s. Yeast clones were grown in AHC and DNA extracted for rescreening by PCR and end rescue by inverse PCR (13). DXYCbl1 is a 282-bp marker produced using the primers CACTATAGTTTTGGCCATAG and GGACGTGTATAATCTGGATG and was derived from the left end of YAC IɛU18. DXYCbl2 is 154-bp product produced from the primers ACATGGACAACTTCGGGAAG and CACACTATTAAGGGACTTCC and was derived from the right end of YAC 42C9. The marker DXCbl1 is a 185-bp product derived from the PCR primers ATCTATCCCTTTTTCTGAGG and CAACATGTTGACAAGTTTGG and was produced by exon trapping the YAC IɛU18. DXYCbl3 is a 194-bp marker amplified using the primers GCCTGAGCAGCATAAAAGAC and TAGGACTAACAAGAGAGGTG and was derived from the YAC 42C9. The sequences of the other markers generated during the walk are available on request.
Southern Hybridization.
For conventional agarose gels, DNA was digested with restriction enzymes and electrophoresed on 0.8% agarose gels. The gels were blotted according to the method of Church and Gilbert (14) and probed overnight by the same method or for shorter periods using Rapid-Hyb (Amersham). For pulsed field gels, DNA embedded in agarose was prepared from kidneys of male C57BL/6 mice and digested with PacI as described (15). Samples were fractionated using a Biometra (Tampa, FL) R23 Rotaphor for 65 h in 0.9% agarose at 13°C in 0.25× TBE (90 mM Tris/64.6 mM boric acid/2.5 mM EDTA, pH 8.3). Gels were alkali-blotted to Oncor Sureblot membrane following the manufacturer’s protocol and hybridized to probes followed by a final wash of 0.2× standard saline citrate (1× standard saline citrate = 0.15 mM sodium chloride/0.015 M sodium citrate, pH 7) at 65°C.
Exon Trapping, cDNA Screening, and Reverse Transcription (RT)–PCR Analysis.
Exons were identified within YACs using the vector λGET (16) and the exon trapping procedure as described (16). An exon discovered using this method was used to probe a 11.5-day postcoitum fetal mouse cDNA library. The resulting two clones were sequenced and each was found to contain an ORF but be truncated at the 5′ end. To obtain the full-length coding sequence, rapid amplification of cDNA ends (17) was performed using cDNA made from testis RNA. The PCR fragments obtained from this experiment were cloned and sequenced and new PCR primers were designed at their extremities. These primers were then used in an RT-PCR using mouse brain cDNA as the template. The resulting fragment was cloned into pBlueScript and sequenced. Both strands of DNA were sequenced using fluorescent chain terminators and analyzed on Applied Biosystems 373 or 377 sequencers. Expression of the Fxy cDNA was analyzed by RT-PCR as described (18) with the following pairs of primers: (i) GAAACACCTGGAGTCGGAGC and GGGCGTGGTCATTTTCCTTC corresponding to nucleotides 126–145 and 1102–1083; (ii) CTTTGAGTGAGCGCTATGAC and TGGGCACGATCATCCAGCTG corresponding to nucleotides 754–773 and 1,447–1,428; and (iii) AGTTCAGCGTGGTCTCCTAC and CCGTACAGTCCAGATGGTCC corresponding to nucleotides 1,345–1,364 and 2,113–2,094. Hprt expression was monitored by PCR with the oligonucleotides GTCAAGGGCATATCCAACAACAAAC and CCTGCTGGATTACATTAAAGCACTG.
RESULTS
Identification of Markers Spanning the Pseudoautosomal Boundary of Mice.
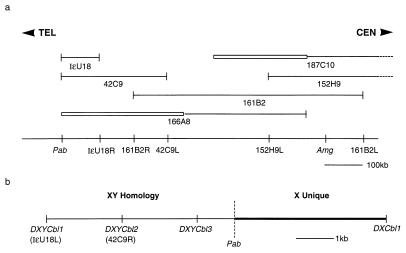
A major obstacle to identification of pseudoautosomal sequences was the paucity of markers in the distal part of the mouse X and Y chromosomes. In initial experiments we attempted to gain access to the mouse PAR using the few markers that had been assigned to this region. However none of these markers identified YACs or P1 clones that mapped to the PAR. Amelogenin (Amg) (19) was the closest defined gene that had been mapped proximal to the boundary on the X chromosome and hence we initiated a genomic walk from this point toward the PAR. Using primers derived from the mouse Amg sequence (19), YACs containing the gene were obtained and the ends rescued by inverse PCR (13). These end fragments were sequenced and primers were designed and tested to produce sequence-tagged site (STSs). Each of these STSs were screened on DNA samples from the European Collaborative Interspecific Backcross (20) or on DNA from mouse/hamster somatic cell hybrids (21) to establish that the YAC end was X-derived before returning to the YAC library for rescreening. At the outset there was no means of knowing the orientation so the walk was bidirectional. The contig developed by this method covers ≈2.5 Mb and the portion of the walk from Amg to the PAR is shown in Fig. 1a. At the end-point of this walk two YAC end STSs (DXYCbl1 and DXYCbl2) mapped to the distal X and also to the Y chromosome and were thus candidates for localization to the PAR (Fig. 1b).
Figure 1.
Physical map of the distal X chromosome and pseudoautosomal boundary region of the mouse. (a) Overlapping YAC clones covering the region between Amg and the pseudoautosomal boundary (Pab). Boxed regions of YACs indicate the presence of non-X chromosomal DNA. The STSs derived from these YACs and their relative distances are indicated below the line. The suffix L or R on the STS name indicates the left or right arm of the YAC from which the STS is derived. The dotted lines on the YACs 187C10 and 152H9 indicate that they extend beyond the map. TEL and CEN indicate telomeric and centromeric directions, respectively. (b) A high-resolution map of the pseudoautosomal boundary showing the positions of the flanking STSs.
The PCR primers used to amplify DXYCbl1 yielded an equivalent sized band in C57BL/6 (B6) and Mus spretus DNA but use of a somatic cell hybrid panel (21) mapped this marker to both the X and Y chromosomes. DXYCbl2, which is located 2 kb proximal to DXYCbl1 is variant between the two species in that the PCR produces a band of 1 kb from B6 DNA but not from M. spretus DNA and could therefore be used to provide a more definitive map location using European Collaborative Interspecific Backcross (20). In 33 European Collaborative Interspecific Backcross DNA samples of animals derived from the F1 backcross to M. spretus (20) no recombinants were found between DXYCbl2 and DXMit12 or DXYMov15, placing it in a very distal X chromosome location.
Using males produced by the F1 female backcrossed to B6, it is possible to distinguish between X-unique and X/Y map locations. The F1 mother has a M. spretus derived X chromosome and a B6-derived X chromosome. Sons of this female inherit X-specific alleles derived from either one species or the other in a hemizygous fashion. Therefore markers such as DXMit12 are positive for the B6 band in only half of the males produced. However, if the marker is also on the Y chromosome, such as DXYMov15, all of the males are positive because they all possess a B6-derived Y chromosome. In 12 males selected from this cross, 5 were positive for the B6-derived DXMit12 band but all 12 were positive for DXYCbl2.
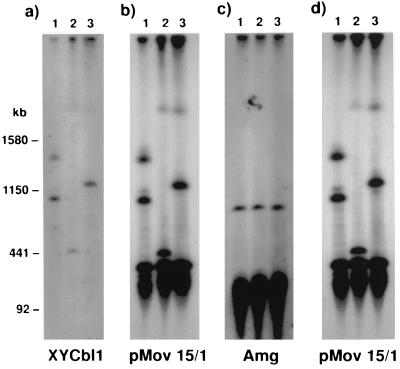
Although these STSs clearly map to the distal portion of the X chromosome this analysis does not allow mapping on the Y chromosome. However the marker DXYCbl1 hybridizes in both males and females to a variable length PacI fragment of known pseudoautosomal origin (22, 23) that cohybridizes with the probe pMov15.1 (Fig. 2). Therefore this analysis demonstrates a physical proximity to known pseudoautosomal markers on both the X and Y chromosomes.
Figure 2.
Physical linkage of DXYCbl1 to a known pseudoautosomal locus. Southern analysis of three C57BL/6 male DNAs cut with PacI and hybridized with DXYCbl1, pMov15/1 and Amg. (a) A blot probed with DXYCbl1 then reprobed in b with pMov15/1 showing the same bands hybridizing. The smallest allele shown in lane two is 500 kb. The lower hybridizing bands in b are derived from other loci containing the macrosatellite repeat including a 400-kb band that is thought to be from chromosome 9. (c) A similar blot probed with Amg showing specific hybridization at 800 kb. The lower smear is nonspecific hybridization to the main bulk of DNA. (d) The same blot shown in c rehybridized with pMov15/1 demonstrating that different bands are identified by this probe. Size markers are Saccharomyces cerevisiae chromosomes (strain YPH148).
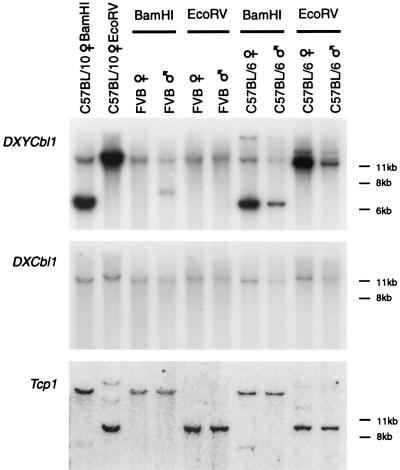
The fact that the walk began from an X-specific location and one extreme of the walk yielded pseudoautosomal markers means that the pseudoautosomal boundary must have been crossed. The opposite ends of the YACs from which DXYCbl1 and DXYCbl2 were derived both map specifically to the X chromosome (Fig. 1a), therefore the search for the boundary could be confined to these YACs. During the course of an experiment to obtain expressed sequences from this region (described below), a DNA marker was rescued that was found to be only 7 kb away from DXYCbl2 and that mapped exclusively to the X chromosome as determined by analysis of the somatic-cell hybrid panel. The X-specific nature of this locus, DXCbl1, is also readily visible by comparing the dosage of hybridizing bands in male and female DNAs (Fig. 3). When the same blot is stripped and probed with DXYCbl1, the identical X-linked band hybridizes in lanes digested with BamHI but gives an additional strong band that is clearly present in multiple copies.
Figure 3.
Southern analysis of the region surrounding the pseudoautosomal boundary. Male and female DNAs from mice of the FVB and C57BL/6 strains and from a female C57BL/10 mouse digested with BamHI and EcoRV were analyzed by Southern blotting and probed with DNA fragments derived from either side of the pseudoautosomal boundary and from an autosomal gene as a loading control. (Middle) A blot probed with DXCbl1 shows that this STS is X-unique and gives a 2:1 ratio of hybridization intensity in female and male lanes. (Top) The same blot stripped and reprobed with DXYCbl1 showing hybridization to the same X-linked band as DXCbl1 in the BamHI cut lanes and an additional, more intense, band at 6.5 kb derived from the pseudoautosomal repeats in the C57BL/10 and C57BL/6 lanes. The repeat band appears to be absent in the FVB female and of an altered allele size in the FVB male. The single 12-kb repeat band in the C57BL/10 female demonstrates that the repeat unit must be at least this size and is probably larger because there are no junction fragments. (Bottom) The same blot stripped and reprobed with a fragment of the autosomal gene Tcp-1 (31) to show equivalence of loading between male and female tracks.
The Mouse PAR Contains a Variable Copy Number Repeat.
The pseudoautosomal probes DXYCbl1, -2, and -3 all hybridize in both male and female DNAs to a repeat band that is usually of greater intensity than the single copy X-specific band (Fig. 3 and data not shown). In a series of digests with a variety of restriction enzymes the repeat band is usually found to be a single very intense band. In a small number of cases, bands of a weaker intensity are also found at different molecular weight to the main band. This suggests that the sequence is remarkably similar between the individual units that comprise this repeat element. The basic repeat unit must be longer than 12 kb because the band is approximately that size in EcoRV-digested DNA without any indications of junction fragments that might suggest a single restriction site within the repeat unit. The complement of repeat units in each individual can be estimated by comparing the intensity of the repeat band against the internal X chromosome control band in DNA cut with BamHI. It is assumed that the female X band is derived from two single copies and the male from one single copy. We estimate, by PhosphorImager analysis, that the female C57BL/10 mouse (Fig. 3) has ≈18 copies. A male of the same strain was found to have ≈24 copies (data not shown). It is clear from the other tracks that the number of repeat units is variable, not sex-specific and in some cases (FVB female) the repeat unit is apparently absent. When the same probe, DXYCbl1, is hybridized to DNA cut with PacI and separated on pulsed field gels, single bands are identified that cohybridize to the known PAR probe pMov15.1 (Fig. 2) (22, 23). If these data are taken in conjunction with the variability of the repeat copy number it appears highly likely that the repeat bands are derived from a tandem repeat array within the PAR that is directly adjacent to the boundary.
A RING Finger Gene Spans the Pseudoautosomal Boundary on the X Chromosome.
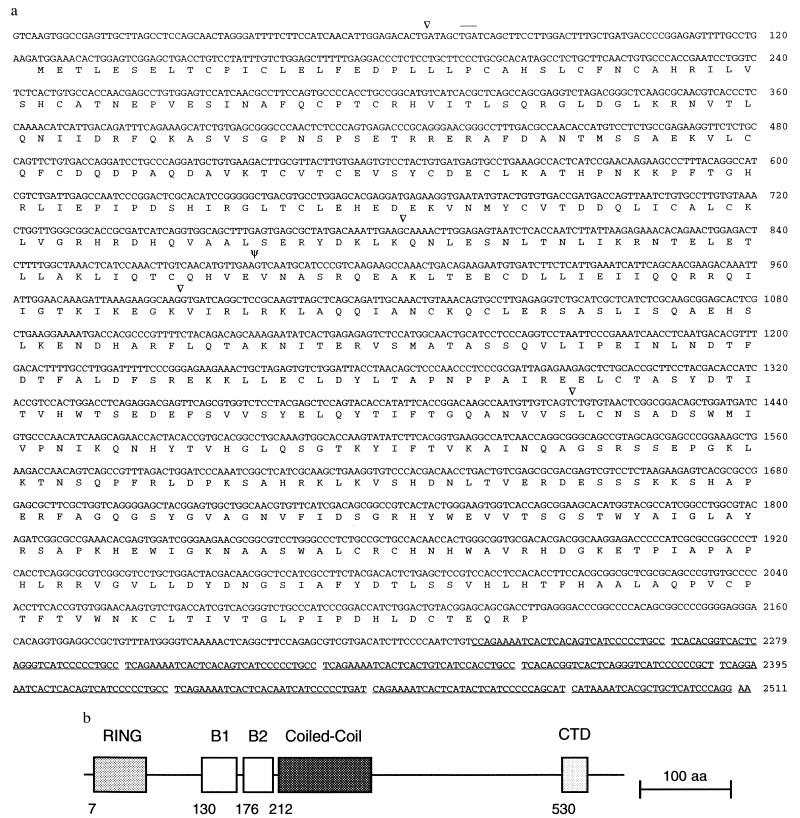
We searched for genes close to the boundary on the X chromosome by exon trapping the YAC IɛU18 from which the YAC-end STS DXYCbl1 was derived (Fig. 1a). An exon was discovered using this method that corresponds to the previously described probe DXCbl1 and is located 7 kb proximal to the pseudoautosomal boundary on the X chromosome. Screening of cDNA libraries and the rapid amplification of cDNA ends procedure (17) was used to isolate cDNA, which was subsequently sequenced (Fig. 4). An in-frame termination codon is present upstream of the putative initiating methionine indicating that we have isolated the entire ORF. The gene, Fxy (ring finger on X and Y), encodes a novel 667-amino acid protein with a RING finger domain (24) near the amino terminus (Fig. 4). Fxy is transcribed at a low level in every tissue tested to date as assayed by RT-PCR (Fig. 5) but the mRNA is not readily detectable by northern blotting. The 3′ rapid amplification of cDNA ends fragment produced contains no polyA tail but 20 bp after the stop codon is a series of tandem repeats that are highly homologous to the macrosatellite repeat that constitutes the DXYMov15 locus (25, 26), located in the pseudoautosomal region.
Figure 4.
Structure of Fxy. (a) DNA and encoded protein sequence of the Fxy gene. Introns are indicated by arrowheads and ψ indicates the intron that contains the pseudoautosomal boundary. Not all the positions of introns 3′ to the boundary have been defined. The DXYMov15 repeats in the 3′-untranslated region of the gene are underlined. An in-frame termination codon present upstream of the putative translation initiating methionine is overlined. (b) Schematic representation of the protein encoded by Fxy. Structural features associated with RING finger proteins (24) are indicated. (Bar = 100 amino acids.)
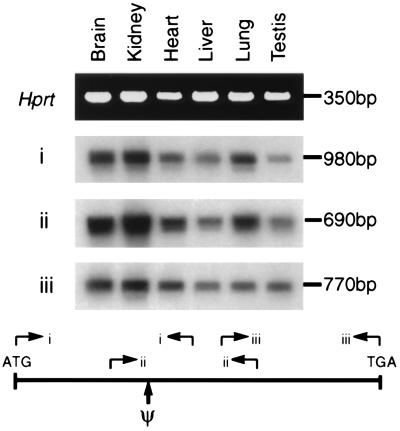
Figure 5.
Expression of the Fxy gene. PCR primer sets (i, ii, and iii) derived from the sequence of Fxy, or as a control Hprt, were used to amplify cDNA made from the indicated tissues. For i, ii, and iii, the products were electrophoresed on an agarose gel, blotted to a membrane, and hybridized with Fxy cDNA. For Hprt a photograph of an ethidium bromide stained gel is shown. Ψ indicates the position of the pseudoautosomal boundary. Multiple introns are present between all the primer pairs and no product is amplified from genomic DNA. The sequences of the primers are described in Materials and Methods.
The gene is oriented in a 5′-3′: centromere-telomere fashion. The exon originally found in the exon trapping experiment at DXCbl1 is within an X-specific region of the chromosome (Fig. 3) as are two more 5′ exons (Fig. 5). However, the remaining 3′ exons all lie within the PAR. Analysis by both PCR and Southern hybridization demonstrated that the exon located 3′ to DXCbl1 is located within the repeat located at the pseudoautosomal boundary. Thus three of the exons of the Fxy gene are located on the X chromosome whereas the 3′ exons of the gene are located on both X and Y chromosomes. If the entire cDNA is used as a probe on the pulsed field gel blot shown in Fig. 2, the same variable PacI fragments are identified as those that hybridize with pMov15.1 and DXYCbl1 (data not shown). Therefore the gene lies wholly within the highly variable PacI fragment that spans the boundary of the X chromosome and occupies a region of the PAR that contains a series of large repeats described above, the macrosatellite repeats of the DXYMov15 locus (25, 26) (also located in the 3′ untranslated region of the Fxy gene; Fig. 4) and a region of telomere-derived (TTAGGG)n simple sequence repeats (22, 23). To confirm that the whole of the gene is transcribed as a unit rather than as two separate genes on either side of the boundary, we performed RT-PCR with primers scattered through the gene (Fig. 5). This showed that the gene is apparently ubiquitously expressed and is transcribed across the pseudoautosomal boundary.
DISCUSSION
The pseudoautosomal boundary has unique properties in mammalian genomes. It is the point of divergence between the pseudoautosomal region, which is identical on the X and Y chromosomes, and X and Y specific sequences. Hence it might be expected that certain constraints would have to be placed on chromosome and gene organization in this vicinity. The only pseudoautosomal boundary that has been characterized previously in any detail is that of primates (27, 28). Here we describe the organization of the X-unique and PAR sequences flanking the pseudoautosomal boundary in the mouse. The PAR sequences immediately adjacent to the boundary are shared by a variable number of large, highly conserved repeating units that all map to this region (22, 23). The variation in the copy number of the repeat could be explained by recombination slippage and unequal exchange. This explanation is supported by the finding that PacI restriction fragments hybridizing to the pseudoautosomal probes pMov15/1, PAR4 (22, 23), and now DXYCbl1 show large variations in size between the DNAs of individual, genetically identical, and inbred mice. We have shown that the precise pseudoautosomal boundary on the X is located at the junction of single copy X-unique DNA and one of the pseudoautosomal variable copy number repeat units in C57BL/6 mice (unpublished work). This has complicated the molecular analysis of the region and the full structure of the boundary on the Y chromosomes remains to be elucidated. However, because the copy number of the repeat appears to vary in almost every individual, the position of the boundary may not be at exactly the same place in every male mouse but may vary according to the number of repeats on each of the sex chromosomes.
We have shown that the pseudoautosomal boundary on the X chromosome is spanned by a transcribed gene, Fxy, encoding a RING finger protein. The three most 5′ exons of the gene including a significant proportion of the coding potential of the gene are X-unique. However the 3′ portion of the gene is present both on the X and Y chromosomes and is included within the repeat present in the PAR. The 3′ exons of the gene are therefore present in multiple copies on the X and Y (Fig. 6). We have not found a transcription unit from the Y chromosome that includes the 3′ exons of Fxy. However, the Y chromosome certainly lacks the 5′ end of this gene that includes the exons that encode the RING finger domain. The Fxy gene is of unknown function but its closest homologue is the gene Rpt-1, which is a signal transduction molecule involved in the regulation of the interleukin 2 receptor (29). The expression pattern of Fxy also gives no clues to its function. The low level of expression in a range of tissues could imply that it has a housekeeping role in all cells or it could mean that the major functional site of expression remains to be determined.
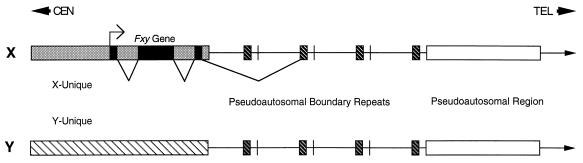
Figure 6.
Schematic representation of the mouse pseudoautosomal boundary region. The three exons at the 5′ end of the gene are X-unique (solid area) but the 3′ region of the gene (dark hatched area) is present on the X and Y chromosomes and within a repeat region just distal to the boundary. Note that for simplicity the 3′ end of the gene is shown as a single block whereas it is actually interrupted by several introns and only four of the variable number of pseudoautosomal repeats (22, 23) are shown. TEL and CEN indicate telomeric and centromeric directions, respectively.
The only other pseudoautosomal boundary that has been described in detail is also spanned by a gene. The human boundary is spanned by the PBDX gene that encodes the XG blood group antigen (30). The orientation of PBDX is 5′-3′: telomere-centromere, which is opposite to the orientation of Fxy across the mouse pseudoautosomal boundary. Therefore both humans and mice have a gene that is intact on the X chromosome but is truncated on the Y chromosome—the mouse Y lacks the 5′ end of Fxy and the human Y the 3′ end of PBDX. It is probable that these truncated versions of the genes are without function and they would therefore not be under selective pressure. Mutations in the Y copy would have no effect on the net functional amount of Fxy protein in a given individual nor would pairing and segregation of the mutant Y chromosome be affected. However, mutation could lead to the suppression of recombination between the X and Y chromosomes in the region proximal to the new mutation because any recombination would transfer the mutation to the functional X copy. Consequently animals inheriting the new mutation in Fxy on their X chromosomes would be expected to suffer deleterious consequences perhaps reducing reproductive fitness. Isolated from recombination, this region of the PAR on the Y chromosome would be susceptible to any number of further mutations without functional consequences apart from a slight shortening of the PAR.
It has been suggested that the PAR in mammals is evolving by the addition of autosomal segments to the PAR together with progressive loss of material from the Y chromosome PAR (12). In this “addition-attrition” hypothesis the site of attrition is the point at which the homologous PAR diverges into X- and Y-specific sequences, i.e., the pseudoautosomal boundary. The action of attrition is part of the ongoing process that, it is suggested, has degraded the Y PAR since the origin of the sex chromosomes from a pair of homologous autosomes (12). We propose that the presence of truncated genes at the pseudoautosomal boundary of the Y chromosome of both humans and mice reveals the process of attrition at work. If this is correct then it seems possible, due to this process, that the Fxy gene might be fully pseudoautosomal, or have been rendered completely X-unique, in other mammalian species.
Acknowledgments
This work was supported by the Cancer Research Campaign and the Medical Research Council. The authors thank the Medical Research Council HGMP staff for supplying DNA samples and YAC clones. S.P. was the recipient of an Medical Research Council Training Fellowship, D.K. is a fellow of the Lister Institute of Preventive Medicine.
ABBREVIATIONS
- RT-PCR
reverse transcription–PCR
- PAR
pseudoautosomal region
- YAC
yeast artificial chromosome
- Amg
Amelogenin
- STS
sequence-tagged site
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF026565).
References
- 1.Ohno S. Sex Chromosomes and Sex-Linked Genes. Berlin: Springer; 1967. [Google Scholar]
- 2.Burgoyne P S. Hum Genet. 1982;61:85–90. doi: 10.1007/BF00274192. [DOI] [PubMed] [Google Scholar]
- 3.Ellis N A, Goodfellow P N. Trends Genet. 1989;5:406–410. doi: 10.1016/0168-9525(89)90199-6. [DOI] [PubMed] [Google Scholar]
- 4.Rouyer F, Simmler M C, Johnsson C, Vergnaud G, Cooke H J, Weissenbach J. Nature (London) 1986;319:291–295. doi: 10.1038/319291a0. [DOI] [PubMed] [Google Scholar]
- 5.Goodfellow P J, Pym B, Mohandas T, Shapiro L. Am J Hum Genet. 1984;36:777–782. [PMC free article] [PubMed] [Google Scholar]
- 6.Ellison J, Passage M, Yu L C, Yen P, Mohandas T K, Shapiro L. Somat Cell Mol Genet. 1992;18:259–268. doi: 10.1007/BF01233862. [DOI] [PubMed] [Google Scholar]
- 7.Slim R, Levilliers J, Ludecke H J, Claussen U, Nguyen V C, Gough N M, Horsthemke B, Petit C. Genomics. 1993;16:26–33. doi: 10.1006/geno.1993.1135. [DOI] [PubMed] [Google Scholar]
- 8.Salido E C, Li X M, Yen P H, Martin N, Mohandas T K, Shapiro L J. Nat Genet. 1996;13:83–86. doi: 10.1038/ng0596-83. [DOI] [PubMed] [Google Scholar]
- 9.Disteche C M, Brannan C I, Larsen A, Adler D A, Schorderet D F, Gearing D, Copeland N G, Jenkins N A, Park L S. Nat Genet. 1992;1:333–336. doi: 10.1038/ng0892-333. [DOI] [PubMed] [Google Scholar]
- 10.Miyajima I, Levitt L, Hara T, Bedell M A, Copeland N G, Jenkins N A, Miyajima A. Blood. 1995;85:1246–1253. [PubMed] [Google Scholar]
- 11.Ferrero G B, Franco B, Roth E J, Firulli B A, Borsani G, Delmas, Mata J, Weissenbach J, Halley G, Schlessinger D, Chinault A C, Zoghbi H Y, Nelson D L, Ballabio A. Hum Mol Genet. 1995;4:1821–1827. doi: 10.1093/hmg/4.10.1821. [DOI] [PubMed] [Google Scholar]
- 12.Graves J A M. BioEssays. 1995;17:311–320. doi: 10.1002/bies.950170407. [DOI] [PubMed] [Google Scholar]
- 13.Ochman H, Gerber A S, Hartl D L. Genetics. 1988;120:621–623. doi: 10.1093/genetics/120.3.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Church G, Gilbert W. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kipling D, Cooke H J. Nature (London) 1990;347:400–402. doi: 10.1038/347400a0. [DOI] [PubMed] [Google Scholar]
- 16.Nehls M, Pfeifer D, Boehm T. Oncogene. 1994;9:2169–2175. [PubMed] [Google Scholar]
- 17.Frohman M A, Dush M K, Martin G R. Proc Natl Acad Sci USA. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ashworth A. In: Transcription Factors: A Practical Approach. Latchman D S, editor. Oxford: Oxford Univ. Press; 1993. pp. 125–142. [Google Scholar]
- 19.Lau E C, Simmer J P, Bringas P, Jr, Hsu D D, Hu C C, Zeichner David, M, Thiemann F, Snead M L, Slavkin H C, Fincham A G. Biochem Biophys Res Commun. 1992;188:1253–1260. doi: 10.1016/0006-291x(92)91366-x. [DOI] [PubMed] [Google Scholar]
- 20.The European Interspecific Backcross Collaborative Group. Hum Mol Genet. 1994;3:621–627. [PubMed] [Google Scholar]
- 21.Williamson P, Holt S, Townsend S, Boyd Y. Mamm Genome. 1995;6:426–432. doi: 10.1007/BF00355646. [DOI] [PubMed] [Google Scholar]
- 22.Kipling D, Wilson H E, Thomson E J, Lee M, Perry J, Palmer S, Ashworth A, Cooke H J. Proc Natl Acad Sci USA. 1996;93:171–175. doi: 10.1073/pnas.93.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kipling D, Salido E C, Shapiro L J, Cooke H J. Nat Genet. 1996;13:78–82. doi: 10.1038/ng0596-78. [DOI] [PubMed] [Google Scholar]
- 24.Saurin A J, Borden K L B, Boddy M N, Freemont P S. Trends Biochem Sci. 1996;21:208–214. [PubMed] [Google Scholar]
- 25.Harbers K, Soriano P, Muller U, Jaenisch R. Nature (London) 1986;324:682–685. doi: 10.1038/324682a0. [DOI] [PubMed] [Google Scholar]
- 26.Harbers K, Franke U, Soriano P, Jaenisch R, Muller U. Cytogenet Cell Genet. 1990;53:129–133. doi: 10.1159/000132912. [DOI] [PubMed] [Google Scholar]
- 27.Ellis N A, Goodfellow P J, Pym B, Smith M, Palmer M, Frischauf A M, Goodfellow P N. Nature (London) 1989;337:81–84. doi: 10.1038/337081a0. [DOI] [PubMed] [Google Scholar]
- 28.Ellis N, Yen P, Neiswanger K, Shapiro L J, Goodfellow P N. Cell. 1990;63:977–986. doi: 10.1016/0092-8674(90)90501-5. [DOI] [PubMed] [Google Scholar]
- 29.Patarca R, Freeman G J, Schwartz J, Singh R P, Kong Q T, Murphy E, Anderson Y, Sheng F Y, Singh P, Johnson K A. Proc Natl Acad Sci USA. 1988;85:2733–2737. doi: 10.1073/pnas.85.8.2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ellis N A, Tippett P, Petty A, Reid M, Weller P A, Ye T Z, German J, Goodfellow P N, Thomas S, Banting G. Nat Genet. 1994;8:285–290. doi: 10.1038/ng1194-285. [DOI] [PubMed] [Google Scholar]
- 31.Willison K R, Dudley K, Potter J. Cell. 1986;44:727–738. doi: 10.1016/0092-8674(86)90839-1. [DOI] [PubMed] [Google Scholar]