SUMMARY
The β-barrels found in the outer membranes of prokaryotic and eukaryotic organisms constitute an important functional class of proteins. Here we present solid-state NMR spectra of the bacterial outer membrane protein OmpX in oriented lipid bilayer membranes. We show that OmpX is folded in both glass-supported oriented lipid bilayers and in lipid bicelles that can be magnetically oriented with the membrane plane parallel or perpendicular to the direction of the magnetic field. The presence of resolved peaks in these spectra demonstrates that OmpX undergoes rotational diffusion around an axis perpendicular to the membrane surface. A tightly hydrogen-bonded domain of OmpX resists exchange with D2O for days and is assigned to the transmembrane β-barrel, while peaks at isotropic resonance frequencies that disappear rapidly in D2O are assigned to the extracellular and periplasmic loops. The two-dimensional 1H/15N separated local field spectra of OmpX have several resolved peaks, and agree well with the spectra calculated from the crystal structure of OmpX rotated with the barrel axis nearly parallel (5° tilt) to the direction of the magnetic field. The data indicate that it will be possible to obtain site-specific resonance assignments and to determine the structure, tilt, and rotation of OmpX in membranes using the solid-state NMR methods that are currently being applied to α-helical membrane proteins.
Keywords: OmpX, beta barrel, outer membrane protein, bilayer, bicelle, solid state NMR
INTRODUCTION
The solid-state NMR orientation-dependent frequencies measured for samples of membrane proteins in oriented lipid bilayers provide very high-resolution restraints for protein structure determination and refinement [1, 2]. Measurements made in oriented lipid bilayers have the important advantage that they enable structures to be determined in an environment that closely resembles the cellular membrane, and since the alignment tensor is fixed by the sample geometry, they provide the global orientation of the protein in the membrane. This information can also be used to supplement the data from micelles, to derive detailed structural information about membrane proteins in cases where the same structure is present in the two types of samples [3]. For proteins in oriented lipid bilayers, the two-dimensional 1H/15N separated local field spectra exhibit characteristic wheel-like patterns of resonances - PISA (polarity index slant angle) wheels - that reflect both the protein structure and orientation in the membrane [4, 5]. The direct relationship between spectrum and structure makes it possible to calculate NMR spectra from specific structural models of proteins and provides a method for structure determination [6, 7]. Distinct PISA wheels have been observed experimentally for α-helical membrane proteins [3-7], but have also been predicted for β-stranded proteins [8], and a recent report describing the reconstitution of the transmembrane domain of E. coli OmpA, in magnetically oriented lipid bilayers, indicates that solid-state NMR of β-barrel outer membrane proteins is feasible [9].
The β-barrel outer membrane proteins form a major functional class, with important roles in regulating molecular transport, bacterial pathogenesis, and mitochondrial homeostasis [10], and therefore represent important targets for structural and functional characterization. For example, bacterial pathogens are capable of neutralizing the host defense system by adhering to and entering the host cells and by interfering with the host immune system, and for gram-negative bacteria, these interactions are mediated by outer membrane proteins, which include E. coli OmpX and its homologs in the human pathogens Yersinia, Enterobacter, Klebisiella, and Salmonella. Within this family of virulence-related β-barrel membrane proteins, the membrane-spanning portion of the protein barrel is much better conserved than the extracellular domain, which forms a protruding β-sheet that has been proposed to promote cell adhesion and invasion, and to defend against the host immune system during pathogenesis [10, 11].
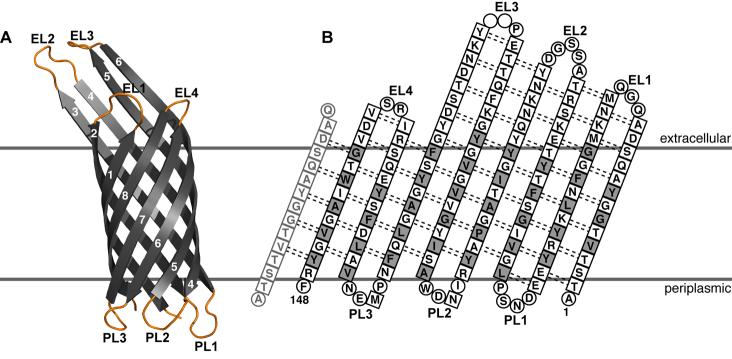
The structures of about twenty β-barrel membrane proteins have been determined, primarily by x-ray crystallography (for reviews see [10, 12-14]), with two also determined by solution NMR in micelles (OmpX, OmpA) [15, 16], and one uniquely determined by solution NMR in micelles (PagP) [17]. The crystal structure of E. coli OmpX [11] consists of eight all-next-neighbor antiparallel β-strands that fold into a cylindrical barrel, with polar residues on the inside and hydrophobic residues on the outside facing the membrane environment (Figure 1). The height of the cylindrical barrel is approximately 32 Å and its diameter 20 Å, and four of the β-strands (β-strands 3-6) extend out of the predicted membrane boundaries to a total height of 50 Å. The strands are connected by three loops on the periplasmic side (PL1-PL3), and four somewhat loser loops on the extracellular side (EL1-EL4). The NMR structure of OmpX in detergent micelles [15] has the same overall features and dimensions as the crystal structure. In the micelle structure, however, the β-strands are about two residues shorter than in the crystal structure, and the loops and turns are less well-defined and exhibit varying degrees of mobility. A recent study on the transmembrane domain of E. coli OmpA found that non-specific physical interactions between the lipid bilayer and the protein are very important for determining protein stability and fold [18]. This argues very strongly in favor of determining the structures of β-barrel membrane proteins in the functional environment of the lipid bilayer, and solid-state NMR spectroscopy is the only method available for this purpose.
Figure 1.
Crystal structure and amino acid sequence of OmpX. (A) Structure of OmpX (PDB file 1QJ8 [11]). The eight β-strands are in gray, and the four extracellular loops (EL) and three periplasmic loops (PL) are in yellow. (B) Amino acid sequence and structural organization of OmpX. Residues in β-strands are in squares, and residues in loops are in circles. Transmembrane residues on the barrel surface are shown in gray. The first strand is shown twice to illustrate the cylindrical connection. Dashed lines indicate hydrogen bonds. Adapted from reference [11].
Although samples of membrane proteins in lipid bilayers are too large for solution NMR structural studies, they are suitable for solid-state NMR experiments where macroscopic alignment of the samples in the magnetic field provides both high-resolution and orientation-dependent restraints for structure determination. Sample alignment can be obtained by assembling planar phospholipids bilayers on silica surfaces or with bicelles - phospholipid bilayers, which orient spontaneously in the magnetic field. Surface-supported bilayers can be prepared using a wide variety of lipids and are the closest analogs to natural membranes for studying membrane proteins, however, they have the disadvantages that it is not easy to maintain a constant level of hydration during the course of NMR experiments, and that the samples cannot be easily manipulated after preparation. Bicelles, in contrast, have the important advantage that the samples are fully hydrated. The use of non-hydrolyzable ether-linked phospholipids contributes significantly to the long-term (months) stability of bicelle samples, and should also benefit planar supported bilayer samples. Both types of samples are extremely valuable for NMR studies of membrane proteins. Here we describe the preparation of solid-state NMR samples of E. coli OmpX in both types of oriented lipid bilayers, and present one-and two-dimensional spectra, which highlight some of the structural features of the protein in membranes.
MATERIALS AND METHODS
Protein expression and purification
The E. coli plasmid pET3b-OmpX was obtained from Georg Schulz. It directs the expression of a mutant form of OmpX, with His100 changed to Asn in the third extracellular loop, which was found to optimize the polar packing contacts in the crystal [11]. The OmpX-bearing plasmid was transformed in E. coli C41(DE3) cells [19] (www.overexpress.com), and the positive clones were grown in M9 minimal media to a cell density of OD600 = 0.7, before induction with IPTG for 3 h. The growth media contained (15NH4)2SO4 as the nitrogen source for 15N labeling. The buffers used in the subsequent purification steps are described in Table 1. Isotopes were from Cambridge (www.isotope.com), lipids were from Avanti Polar Lipids (www.avantilipids.com), and the detergents C8POE and C4E8 were from Anatrace (www.anatrace.com). SDS-PAGE (sodium dodecyl sulfate-polyacrylamide gel electrophoresis) was performed with the Tris-Tricine system [20], and gels were stained with Coomassie Blue.
Table 1.
List of buffers used for protein purification and sample preparation.
| buffer A: (20 mM Tris, pH 8, 2 mM NaN3) |
| buffer AT: (20 mM Tris, pH 8, 2 mM NaN3, 2% w/v Triton X-100) |
| buffer AU: (20 mM Tris, pH 8, 2 mM NaN3, 6 M urea) |
| buffer AP5: (20 mM Tris, pH 8, 2 mM NaN3, 5% w/v C8POE) |
| buffer AP1: (20 mM Tris, pH 8, 2 mM NaN3, 1% w/v C8POE) |
| buffer A6: (20 mM Tris, pH 8; 150 mM 6-O-PC) |
| buffer B: (20 mM Tris, pH 7, 200 mM KCl, 0.1 mM EDTA) |
| buffer BE: (20 mM Tris, pH 7, 200 mM KCl, 0.1 mM EDTA, 1% w/v C4E8) |
| buffer C: (20 mM Tris, pH 9.5) |
| buffer D: (1 mM Tris-HCl, pH 8) |
Protein purification
OmpX was purified as described by Schulz and coworkers [11], with minor modifications. Cells from 1 L of culture were lysed by French Press in 30 mL of buffer A, and the soluble fraction was removed by centrifugation (48,000 × g, 4°C, 30 min). The pellet was re-suspended and mixed in 30 mL of buffer AT for 2 h, followed by centrifugation (48,000 × g, 4°C, 30 min) to remove the soluble fraction. The resulting pellet, enriched in OmpX, was dissolved in buffer AU, and OmpX was purified by ion-exchange chomatography with a NaCl gradient (FF-Q HiPrep-Q column, Amersham). Pure OmpX eluted at 180 mM NaCl, and was concentrated to 2.5 mg/mL.
Samples in glass-supported lipid bilayers
The OmpX solution (10 mL of 2.5 mg/mL in buffer AU) was first diluted by adding 50 mL of buffer AP5, then extensively dialyzed against buffer AP1 followed by buffer BE, and concentrated to 1.5 mg/mL. The lipids, 80 mg of DOPC (di-oleoyl-phosphatidylcholine) plus 20 mg of DOPG (dioleoyl-phosphatidylglycerol), were mixed in chloroform, and the solvent was evaporated under a stream of nitrogen followed by high vacuum for 1 h. The dry lipid mixture was then suspended in 5 mL of buffer B and sonicated to transparency using a Branson sonifier with a micro tip (www.sonifier.com) to form small unilamellar vesicles. For reconstitution, 3 mL of OmpX solution (1.5 mg/mL in buffer BE) were added to 5 mL of the lipid vesicle suspension and mixed by vortexing. The lipid / protein mixture was diluted by the addition of 12 mL of buffer B, rapidly frozen in liquid nitrogen, allowed to thaw at room temperature, and bath-sonicated for 30 sec. The preparation was transferred to a 10,000 molecular weight cutoff dialysis bag (www.spectrapor.com) and dialyzed against four 12 h changes of 4 L of buffer B, followed by two 5 h changes of 4 L of water, all at 4°C.
The reconstituted vesicle preparation was concentrated by ultrafiltration in an Amicon cell with a YM10 membrane (www.millipore.com), and evenly distributed over the surface of fifteen glass cover slides (www.marienfeld-superior.com) with dimensions 11 × 11 × 0.07 mm. After allowing the bulk water to evaporate, the slides were stacked and placed in a sealed chamber together with a saturated ammonium phosphate solution, which provides 93% relative humidity. Oriented bilayers formed after equilibrating the sample in this chamber at 42°C for 12 h. For HD exchange, the stacked sample was incubated in a chamber where H2O had been replaced with D2O, at 42°C for several hours.
The samples contained 4.5 mg of uniformly 15N-labeled OmpX, 80 mg of DOPC, and 20 mg of DOPG, corresponding to a protein to lipid molar ratio of 1/400. Before insertion into the coil of the NMR probe, the stack of glass slides was wrapped in a thin layer of parafilm and then placed in thin polyethylene tubing, which was heat sealed at both ends to maintain sample hydration during the NMR experiments.
Samples in lipid bicelles
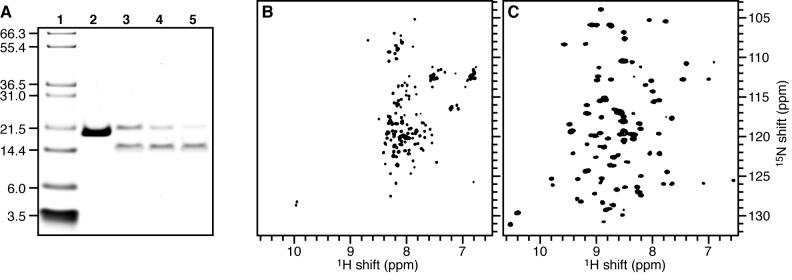
The purified OmpX solution (3.2 mL of 2.5 mg/mL in buffer AU) was dialyzed against water to remove Urea and salts, and pure OmpX was lyophilized to obtain a white powder. For the bicelle samples, 8 mg of lyophilized OmpX were dissolved in 1 mL of 100 mM SDS in water, and added to small unilamellar vesicles, prepared by sonicating 46.5 mg of 14-O-PC (1,2-O-ditetradecyl-sn-glycero-3-phosphocholine) in 20 mL of buffer C, as described above. The protein / detergent / lipid solution was incubated for at least 12 h at 40°C, and folding was monitored by SDS-PAGE, where the appearance of a band with lower apparent molecular weight corresponds to folded OmpX (Figure 2A). After refolding, SDS was removed by dialysis against several 3-12 h changes of 1 L of buffer C, followed by buffer D.
Figure 2.
Refolding of OmpX in lipids. (A) SDS-PAGE monitoring OmpX refolding in 14-O-PC small unilamellar vesicles. (Lane 1) Molecular weight markers. (Lane 2) OmpX dissolved in 6 M urea. (Lanes 3-5) OmpX dissolved in 100 mM SDS and added to 14-O-PC vesicles to initiate protein refolding; samples were collected: (lane 3) immediately after start of refolding reaction, (lane 4) after 2 h incubation, or (lane 5) after 12 h incubation. Unfolded and folded OmpX migrate at molecular weights of 20 kD and 16 kD, respectively. All samples were loaded without boiling. (B, C) Solution NMR 1H/15N HSQC spectra of uniformly 15N labeled OmpX in (B) 500 mM SDS and (C) 150 mM 6-O-PC.
The sample was concentrated by ultrafiltration in an Amicon cell using a YM10 membrane, to a final volume of 100 μL. For the unoriented sample, this concentrated OmpX / 14-O-PC preparation was transferred directly to a flat-bottomed 5 mm tube (www.newera-spectro.com) that was sealed with a rubber cap. For bicelle preparation, 9.5 mg of 6-O-PC in 100 μL of buffer D were added to the OmpX / 14-O-PC preparation, and bicelles were formed by repeated freeze-thawing as described previously [21, 22], then transferred to a flat-bottomed 5 mm tube and sealed with a rubber cap.
The bicelle samples contained 8 mg OmpX, 46.5 mg 14-O-PC, 9.5 mg 6-O-PC, in 200 μL of buffer D. The molar ratio (q) of long to short chain phospholipids was 3.2, and the protein to lipid molar ratio was 1/190. Bicelles were flipped by adding 5 μL of 100 mM YbCl3 (www.sigma.com) directly to the NMR tube pre-cooled at 4°C, mixing thoroughly, and re-sealing the tube. HD exchange experiments were carried out by first reducing the sample volume to 100 μL under a stream of N2 gas, then adding 100 μl of D2O, and repeating this process 4 times.
Samples in micelles
For samples in 6-O-PC (1,2-O-dihexyl-sn-glycero-3-phosphocholine), refolding of OmpX was obtained by slow dropwise addition of 4 mg of OmpX, in 20 μL of buffer AU, to 200 μL of buffer A6, followed by dialysis against 4 L of buffer D at 4°C to remove urea. To minimize the loss of 6-O-PC, two 20 min dialysis steps were carried out using a 1,000 molecular weight cutoff membrane to achieve a final urea concentration of 1.5 nM. Some protein precipitation was observed during this process, and the precipitated protein was removed by centrifugation. The final protein concentration in the NMR sample was 0.2 mM OmpX in 500 μL of 150 mM 6-O-PC, 1 mM Tris-HCl, pH 8, and 5% D2O. For samples in SDS, urea was removed by dialysis against water and pure OmpX was lyophilized to obtain a white powder. Lyophilized OmpX (4 mg) was dissolved in 500 μL of buffer D supplemented with 500 mM deuterated SDS and 5% D2O, to obtain a 0.5 mM protein sample.
NMR experiments
Solution NMR experiments were performed on a Bruker AVANCE 500 spectrometer at 30° C. The standard FHSQC (fast heteronuclear single quantum correlation) experiment was used for isotropic samples with 1024 points in t2 and 256 in t1 [23]. Solid-state NMR experiments were performed on a Bruker AVANCE 500 spectrometer (www.bruker-biospin.com) equipped with a 500/89 AS Magnex magnet (www.magnex.com), using double-resonance (1H/15N or 1H/31P) probes built at the UC San Diego NIH Resource for Molecular Imaging of Proteins (nmrresource.ucsd.edu), with a 5 mm inner diameter cylindrical coil for the bicelle samples, or a 11 × 11 × 4 mm square coil for the glass-supported bilayer samples. Bicelle samples were allowed to equilibrate in the magnetic field for at least 2 h at 42.5°C before experiments. One-dimensional 15N chemical shift spectra were obtained using single contact CPMOIST [24], and two-dimensional 1H/15N separated local field spectra were obtained with SAMMY [25], with 1H irradiation field strengths of 50-55 kHz, a cross-polarization time of 1 msec, a recycle delay of 8 sec, acquisition times of 5 msec or 10 msec, and SPINAL-16 heteronuclear decoupling during acquisition [26, 27]. For the SAMMY spectra, 250-350 transients were acquired for each of 128 t1 increments. Other parameters were as described [25, 28]. 31P chemical shift spectra were obtained with a single pulse and continuous 1H irradiation (20 kHz field strength) during acquisition to decouple the 1H-31P dipolar interactions. The chemical shifts were referenced to the 1H2O resonance set to its expected position of 4.5 ppm at 42.5°C [29].
Data analysis and calculations
The NMR data were processed using NMRPipe [30], and the spectra were analyzed using Sparky [31]. Solid state NMR spectra were calculated with tensor values and FORTRAN code as described previously [8], from the coordinates of the 1.90Å OmpX crystal structure (PDB file 1QJ8) [11], rotated with the barrel axis nearly parallel to the magnetic field. The spectra were calculated by treating residues in the β-strands as static and residues in the extracellular and periplasmic loops as disordered and mobile. This was obtained by scaling the loops NMR frequencies (F), calculated from the crystal structure, by a factor of 5 around the isotropic chemical shift (119 ppm), to account for the ratio of the full span of 15N chemical shift tensor (∼ 150 ppm) to the isotropic span (∼ 25 ppm), using the formula 119 + (|F − 119| / 5) for F > 119 ppm, or 119 − (|F − 119| / 5) for F < 119 ppm.
RESULTS AND DISCUSSION
SDS-PAGE analysis of the folded state of OmpX
To ascertain that OmpX was of sufficient purity for NMR studies we first examined the purified protein by SDS-PAGE and solution NMR (Figure 2). The gel shows that purified OmpX runs as a single band (Figure 2A, lane 2), and the 1H/15N HSQC spectrum of OmpX in 6-O-PC (Figure 2C) is well-dispersed and very similar to the spectrum previously reported for OmpX in DHPC (1,2-dihexyl-sn-glycero-3-phosphocholine), the ester-linked analog of 6-O-PC [15]. The presence of a single peak for each amino acid residue indicates that the sample is pure and homogeneously folded in 6-O-PC micelles.
SDS-PAGE analysis can be very useful for monitoring folding of β-barrel outer membrane proteins, since the unfolded and folded states give bands with distinct molecular weights [11, 14]. Thus, while purified OmpX in 6 M Urea has an apparent molecular weight near 20 kD (Figure 2A, lane 2), protein folding from SDS solution into 14-O-PC vesicles could be monitored by the appearance, over time, of a band near the calculated molecular weight of the protein (16 kD) and the disappearance of the higher molecular weight band (Figure 2A, lanes 3-5). After incubating the OmpX / SDS / 14-O-PC mixture for 12 hours (Figure 2A, lane 5), the presence of a band near 16 kD indicated folded protein, and most of the intensity corresponding to unfolded protein was gone. The small 1H chemical shift dispersion (∼ 1 ppm) in the HSQC spectrum of OmpX in SDS micelles indicates that the protein is not properly folded in this detergent (Figure 2B), however incubation with phospholipids leads to folding in a short period of time. To further promote protein folding, we used a pH above neutral, since this was previously reported to promote folding of OmpA in DMPC (dimirystoyl-phosphatidylcholine) the ester-linked analog of 14-O-PC [32]. Reconstitution of OmpX in 14-O-PC was the starting point for the preparation of OmpX in bicelles.
OmpX in glass-supported lipid bilayers
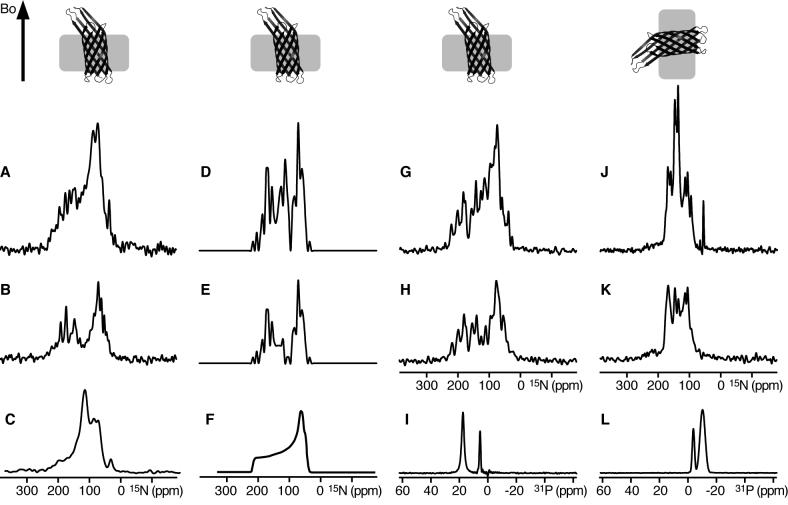
To reconstitute OmpX in phospholipid bilayers we also tested a method that has proved useful for the efficient reconstitution and solid-state NMR sample preparation of other integral membrane proteins [33, 34]. We started from purified OmpX dissolved in the detergent C4E8, since this was used successfully for protein crystallization by Schulz and coworkers [11]. This solution was mixed with pre-formed small unilamellar lipid vesicles, followed by a freeze-thaw cycle and dialysis prior to deposition on glass slides for sample alignment. The resulting 15N chemical shift spectra are shown in Figure 3 (A, B), and are compared with the spectra calculated from the x-ray coordinates of OmpX (Figure 3D, E).
Figure 3.
Solid-state NMR spectra of uniformly 15N labeled OmpX in phospholipid bilayers. The corresponding orientations of OmpX membranes relative to the magnetic field (Bo) are shown at the top. (A, B) Supported DOPC/DOPG lipid bilayers oriented on glass slides with the membrane plane perpendicular to the magnetic field, in (A) H2O, or (B) D2O. (C) OmpX in unoriented 14-O-PC vesicles. (D, E) Spectra calculated from the crystal structure of OmpX rotated with the barrel axis tilted by 5° relative to the direction of the magnetic field (Bo), showing, (D) peaks from all sites, or (E) only peaks from β-strands but not from the loops. (F) Powder pattern calculated for static amide sites. (G-L) Lipid bicelles magnetically oriented with the membrane plane, (G-I) flipped perpendicular to the magnetic field by adding Yb3+, or (J-L) parallel to the magnetic field. Spectra were obtained (G, J) in H2O, or (H, K) in D2O. (I, L) 31P NMR spectra of the oriented bicelle lipids. The intensities of the experimental spectra were set to display a similar signal-to-noise level in the baseline. The intensities of the calculated spectra were set to match those of the experimental spectra.
The spectrum of OmpX in oriented bilayers (Figure 3A) has some discernable resonances, and its overall shape and frequency range are similar to those observed in the spectrum calculated for OmpX oriented with the barrel axis nearly parallel (5°) to the magnetic field (Figure 3D). Notably, it is very different from the spectrum obtained for OmpX in unoriented lipid vesicles (Figure 3C), which is a powder pattern spanning the full range (60-220ppm) of the amide 15N chemical shift interaction, similar to the calculated powder pattern in Figure 3F, except for the additional intensity near the isotropic frequencies (100-130ppm), which reflects the presence of more mobile disordered residues in the sample, and probably corresponds to the extracellular and periplasmic loops of OmpX.
To examine the degree of hydrogen bonding, we exposed the oriented OmpX sample to a D2O atmosphere to exchange labile hydrogens for deuterons. Amide hydrogen exchange rates are useful for identifying residues that are involved in hydrogen bonding. For example, the amide hydrogens in transmembrane helices can have very slow exchange rates due to strong hydrogen bonds in the low dielectric environment of the lipid bilayer, and their 1H-cross-polarized 15N signals can persist for days after exposure to D2O [35]. Faster exchange rates are observed for transmembrane helices that are not tightly hydrogen bonded and are in contact with water because they participate in channel pore formation [36], and for other water-exposed regions of proteins with weaker hydrogen bonded networks. When the OmpX sample was exposed to D2O (Figure 3B), several amide hydrogens exchanged, and their peaks disappeared from the spectrum, while the remaining resonances persisted well after 48 hours, demonstrating the presence of a tight hydrogen bond network, and also confirming the presence of folded protein in the oriented lipid bilayers. Overall, HD exchange reduces resonance overlap and simplifies the spectrum, but the principal effect of HD exchange is to remove the peaks near the isotropic frequency, and near 35 ppm from amino groups of the lysine sidechains and the N-terminus. Taken together with the isotropic intensity observed in the spectrum from unoriented vesicles, this indicates that the majority of exchanged sites correspond to disordered mobile residues in the protein.
The spectrum of OmpX in D2O also demonstrates that a high degree of alignment can be obtained with these samples. This is further confirmed by comparison with the spectra calculated from the crystal structure of OmpX. The spectra in Figures 3D and 3E were obtained by treating residues in the β-strands as structured and static, and those in the extracellular and periplasmic loops as disordered and mobile by scaling their calculated frequencies to the isotropic range. The spectrum showing peaks from all sites (Figure 3D) compares very well with that obtained experimentally for oriented OmpX in H2O (Figure 3A), and when the isotropic resonances from the loops were removed to simulate the effect of HD exchange, the resulting spectrum (Figure 3E) matches extremely well with the experimental spectrum obtained for oriented OmpX in D2O (Figure 3B).
These results indicate that OmpX adopts a similar structure in lipid bilayers as in the crystal, and are consistent with the extracellular and periplasmic loops being more disordered and able to rapidly exchange their amide hydrogens, although this will have to be confirmed with high-resolution structural studies. These one-dimensional spectra are useful for characterizing the dynamics of the protein, and are the basis for higher dimensional NMR experiments and structure determination.
OmpX in lipid bicelles
Bicelles are planar lipid bilayers composed of mixtures of long and short chain phospholipids. Large bicelles (q > 2.5) orient spontaneously in the NMR magnet with the bilayer plane parallel to the direction of the magnetic field, and can be flipped by 90° with the addition of lanthanide ions, to obtain spectra similar to those for planar glass-supported bilayers (reviewed in [37, 38]). Fast uniaxial diffusion around the axis normal to the lipid bilayer results in single-line spectra even for unflipped bicelles [21, 22, 39], and the resulting orientation-dependent frequencies can be used for protein structure determination. Performing experiments at this orientation has the important advantage of reducing the radiofrequency range of the anisotropic interactions, thereby relieving the requirements for high power decoupling, and allowing experiments to be performed at higher magnetic fields. High-resolution multi-dimensional solid-state NMR spectra have been reported for several 15N-labeled helical membrane proteins in magnetically aligned bicelles [21, 22], including spectra for a G-protein coupled receptor [39], and one-dimensional 15N chemical shift spectra have been reported for the transmembrane domain of the E. coli β-barrel OmpA [9].
The spectra in Figure 3 (G-L) show that OmpX can be incorporated in lipid bicelles oriented with the bilayer plane parallel to the field of the NMR magnet (unflipped bicelles; Figure 3J-L), or perpendicular to it (flipped bicelles; Figure 3G-I). The spectrum of OmpX in unflipped bicelles is not a powder pattern but shows discrete peaks from amide sites over a range from about 70 to 160 ppm (Figure 3J), indicating that the protein or protein / bicelle assembly undergo rotational diffusion (> 105 sec−1) around an axis perpendicular to the plane of the lipid bilayer. This is similar to previous observations with helical membrane proteins [21, 22, 39]. The very small trace of residual powder pattern discernable in this spectrum reflects a small amount of OmpX that forms a precipitate and remains unoriented, and might be corrected by experimenting with lower protein concentrations and different lipid compositions.
Flipping the bicelles by 90° yields a very different spectrum spanning a much wider range of 15N chemical shift frequencies (Figure 3G). As expected, discrete peaks can be seen at this orientation, and the spectrum now resembles the spectrum obtained for OmpX in supported lipid bilayers, which are oriented in a similar way. The high degree of uniaxial alignment of both unflipped and flipped bicelles is demonstrated by the 31P NMR spectra of the lipids (Figures 3I, 3L), which display single lines at the expected frequencies corresponding to signals from 14-O-PC (−11 ppm unflipped; 18 ppm flipped) and 6-O-PC (−4 ppm unflipped; 6 ppm flipped).
As observed for OmpX in supported bilayers, the addition of D2O causes several amide hydrogens to exchange for deuterons, with the principal effect of reducing resonance overlap and reducing the intensity of peaks near the isotropic chemical shift frequencies for both the unflipped (Figures 3K) and flipped (Figures 3H) bicelle samples.
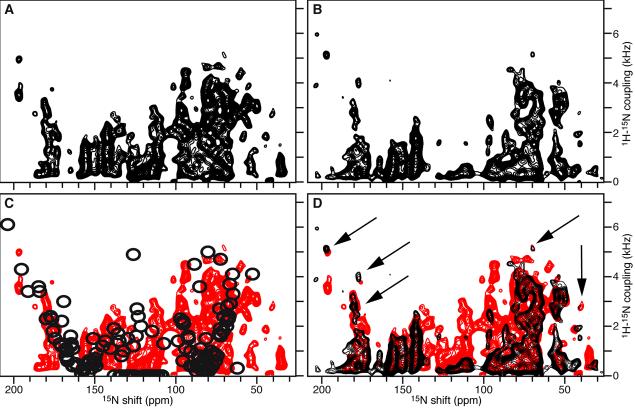
The effect of HD exchange is even more evident in the two-dimensional 1H/15N separated local field spectra obtained for OmpX in flipped bicelles (Figure 4). Comparison of the spectra obtained in H2O (Figure 4A) and D2O (Figure 4B) shows that HD exchange causes several peaks to disappear. Many of the lost peaks are near the isotropic chemical shift and dipolar coupling frequencies, and probably correspond to the extra-membrane loops of the protein, as discussed above. As seen in Figure 4D, the spectrum obtained in H2O (red) overlaps very well with the spectrum obtained after exchange in D2O (black), including for several resolved peaks (arrows). This indicates that the sample is stable over the course of the NMR experiments, and over the course of sample manipulations (HD exchange in this case), allowing reproducible spectroscopic results to be obtained. Both spectra compare well with the spectrum back-calculated from the crystal structure of OmpX (Figure 4C, black), assuming static structured β-strands, and disordered mobile loops with frequencies near the isotropic regions of the 15N chemical shift (∼105-135 ppm) and 1H-15N dipolar coupling (0 kHz) tensors.
Figure 4.
Separated local field 1H/15N SAMMY spectra of uniformly 15N-labeled OmpX in flipped bicelles, obtained in (A) H2O, or (B) D2O. (C) The spectrum obtained in H2O (red) is superimposed with the spectrum calculated from the OmpX crystal structure rotated with the barrel axis 5° to the direction of the magnetic field (black). (D) The spectrum obtained in H2O (red) is superimposed with the spectrum obtained after addition of D2O (black). The arrows point to selected resolved peaks that are unchanged in the two spectra.
The calculated spectra were derived by rotating the coordinates of the 1.90Å crystal structure of OmpX to obtain a 5° tilt of the barrel axis relative to the magnetic field (Figure 5A). The agreement between the spectra obtained in bicelles and those calculated from the crystal structure appears to be quite good, however, a quantitative comparison of the crystal and bicelle structures is premature in the absence of resonance assignments to specific amino acids or to amino acid type. We anticipate that structural comparisons will be possible once the spectra from selectively labeled samples are obtained. Furthermore, although a 5° tilt reproduced the overall features of the one- and two-dimensional experimental NMR spectra better than a 0° or a 10° tilt, we note that a detailed rotational analysis and spectrum back-calculation was not carried out at this early stage of the research, and therefore, that the data are not sufficient to definitively determine the barrel tilt. Nevertheless, it is interesting to note that studies of β-barrel membrane proteins in elliptical micelles or membranes, by molecular dynamics simulations [40], infrared spectroscopy [41], and solution NMR paramagnetic relaxation enhancements [42], indicate that the barrels adopt tilted transmembrane orientations, and that the tilts are dictated by the hydrophobic thickness of the lipid bilayer, as observed for helical membrane proteins [43-46]. The spectra presented in Figures 3 and 4 indicate that it will be possible to determine the tilt and structure of OmpX in membranes.
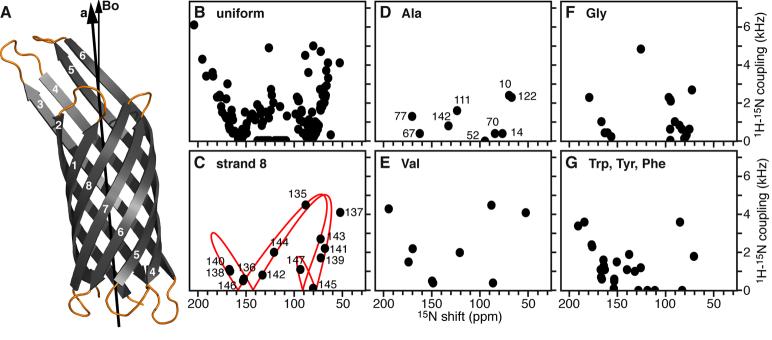
Figure 5.
Separated local field 1H/15N spectra calculated from the coordinates of the 1.9 Å crystal structure of OmpX rotated with the β-barrel axis tilted 5° from the direction of the magnetic field (Bo). (A) Tilted structure of OmpX (PDB file 1QJ8), with loops shown in yellow. (B-G) Spectra calculated for (B) uniformly 15N-labeled OmpX, and (D-G) selectively 15N-labeled OmpX. (C) The peaks from the 8th β-strand fit on the twisted PISA wheel of an ideal β-strand with a tilt of 40° (red line).
Three-dimensional spectroscopy and selectively labeled samples will be required to resolve and assign the spectrum of OmpX in oriented lipid bilayers. The calculated spectra in Figure 5 suggests that most of the resonances will fall on a common twisted PISA wheel pattern, as predicted for ideal β-strands crossing the membrane with angles between 40° and 50° [8]. The separation of even- and odd-numbered resonances on opposite wings of the twisted PISA wheel is apparent when the peaks from the eighth β-strand of OmpX are examined separately (Figure 5C), suggesting that these types of spectra will provide valuable information about transmembrane protein orientation even before complete structure determination. Furthermore, the spectra calculated for selectively 15N-labeled OmpX indicate that it will be possible to resolve individual peaks and measure frequencies for structure determination (Figure 5D-G). In the case of 15N-Ala labeling, for example, the calculated spectrum displays excellent resolution. OmpX has 10 alanines, and since at least 8 or 9 are in β-strands (Ala1 is the N-terminus) the characteristic twisted PISA wheel resonance pattern is readily seen in the spectrum.
CONCLUSIONS
In conclusion, we have demonstrated the preparation of highly oriented samples of 15N-labeled OmpX in both glass-supported lipid bilayers, and in lipid bicelles that can be magnetically oriented either parallel or perpendicular to the magnetic field. The observation of discrete peaks in the spectrum from unflipped bicelles demonstrates that OmpX, or OmpX-containing bicelles, undergo rotational diffusion around an axis perpendicular to the membrane surface, and extends the types of samples that can be used for NMR structural analysis of OmpX. The observation of a strongly hydrogen bonded domain that resists exchange with D2O for days, indicates that the OmpX β-barrel is folded in the lipid bilayer membrane, while the coincidence of peaks at isotropic frequencies with peaks that disappear upon HD exchange, indicates that these correspond to disordered mobile residues in the extracellular and periplasmic loops of OmpX.
Finally, the observation of several resolved peaks in the separated local field spectra of OmpX in flipped bicelles indicates that site-specific resonance assignments will be possible through the use of selectively labeled samples and higher dimensional spectroscopy. The experimental spectra agree well with the spectra calculated from the 1.90Å crystal structure of OmpX with the barrel axis tilted 5° from the magnetic field, suggesting that OmpX may be tilted in the membrane. While a definitive determination of the barrel transmembrane tilt is premature at this stage, the data presented in this report indicate that it will be possible to determine the structure, tilt, and rotation of OmpX in the membrane, using the solid-state NMR methods that are currently being applied to α-helical membrane proteins.
ACKNOWLEDGEMENTS
We thank Georg Schulz for generously providing the plasmid and methods for OmpX expression and purification. We thank Anna DeAngelis and Jinghua Yu, for their assistance with sample preparation and NMR spectroscopy. This work was supported by the NIH (R21GM075917). It utilized the NIH-supported Resource for NMR Molecular Imaging of Proteins (P41EB002031) and the NIH-supported Burnham Institute NMR Facility (P30CA030199).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Cross TA, Quine JR. Protein structure in anisotropic environments: development of orientational constraints. Concepts in Magn. Reson. 2000;12:55–70. [Google Scholar]
- 2.Opella SJ, Marassi FM. Structure determination of membrane proteins by NMR spectroscopy. Chem. Rev. 2004;104:3587–3606. doi: 10.1021/cr0304121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Franzin CM, Teriete P, Marassi FM. Structural similarity of a membrane protein in micelles and membranes. J. Am. Chem. Soc. 2007;129:8078–8079. doi: 10.1021/ja0728371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marassi FM, Opella SJ. A solid-state NMR index of helical membrane protein structure and topology. J. Magn. Reson. 2000;144:150–155. doi: 10.1006/jmre.2000.2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang J, Denny J, Tian C, Kim S, Mo Y, Kovacs F, Song Z, Nishimura K, Gan Z, Fu R, Quine JR, Cross TA. Imaging membrane protein helical wheels. J. Magn. Reson. 2000;144:162–167. doi: 10.1006/jmre.2000.2037. [DOI] [PubMed] [Google Scholar]
- 6.Marassi FM, Opella SJ. Simultaneous assignment and structure determination of a membrane protein from NMR orientational restraints. Protein Sci. 2003;12:403–411. doi: 10.1110/ps.0211503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Asbury T, Quine JR, Achuthan S, Hu J, Chapman MS, Cross TA, Bertram R. PIPATH: an optimized algorithm for generating alpha-helical structures from PISEMA data. J. Magn. Reson. 2006;183:87–95. doi: 10.1016/j.jmr.2006.07.020. [DOI] [PubMed] [Google Scholar]
- 8.Marassi FM. A simple approach to membrane protein secondary structure and topology based on NMR spectroscopy. Biophys. J. 2001;80:994–1003. doi: 10.1016/S0006-3495(01)76078-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Triba MN, Zoonens M, Popot JL, Devaux PF, Warschawski DE. Reconstitution and alignment by a magnetic field of a beta-barrel membrane protein in bicelles. Eur. Biophys. J. 2006;35:268–275. doi: 10.1007/s00249-005-0014-x. [DOI] [PubMed] [Google Scholar]
- 10.Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 2003;67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vogt J, Schulz GE. The structure of the outer membrane protein OmpX from Escherichia coli reveals possible mechanisms of virulence. Structure. 1999;7:1301–1309. doi: 10.1016/s0969-2126(00)80063-5. [DOI] [PubMed] [Google Scholar]
- 12.Wimley WC. The versatile beta-barrel membrane protein. Curr. Opin. Struct. Biol. 2003;13:404–411. doi: 10.1016/s0959-440x(03)00099-x. [DOI] [PubMed] [Google Scholar]
- 13.Schulz GE. The structure of bacterial outer membrane proteins. Biochim. Biophys. Acta. 2002;1565:308–317. doi: 10.1016/s0005-2736(02)00577-1. [DOI] [PubMed] [Google Scholar]
- 14.Tamm LK, Hong H, Liang B. Folding and assembly of beta-barrel membrane proteins. Biochim. Biophys. Acta. 2004;1666:250–263. doi: 10.1016/j.bbamem.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 15.Fernandez C, Hilty C, Wider G, Guntert P, Wuthrich K. NMR structure of the integral membrane protein OmpX. J. Mol. Biol. 2004;336:1211–1221. doi: 10.1016/j.jmb.2003.09.014. [DOI] [PubMed] [Google Scholar]
- 16.Arora A, Abildgaard F, Bushweller JH, Tamm LK. Structure of outer membrane protein A transmembrane domain by NMR spectroscopy. Nat. Struct. Biol. 2001;8:334–338. doi: 10.1038/86214. [DOI] [PubMed] [Google Scholar]
- 17.Hwang PM, Choy WY, Lo EI, Chen L, Forman-Kay JD, Raetz CR, Prive GG, Bishop RE, Kay LE. Solution structure and dynamics of the outer membrane enzyme PagP by NMR. Proc. Natl. Acad. Sci. U. S. A. 2002;99:13560–13565. doi: 10.1073/pnas.212344499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hong H, Tamm LK. Elastic coupling of integral membrane protein stability to lipid bilayer forces. Proc. Natl. Acad. Sci. U. S. A. 2004;101:4065–4070. doi: 10.1073/pnas.0400358101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miroux B, Walker JE. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 1996;260:289–298. doi: 10.1006/jmbi.1996.0399. [DOI] [PubMed] [Google Scholar]
- 20.Schagger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 21.De Angelis AA, Jones DH, Grant CV, Park SH, Mesleh MF, Opella SJ. NMR experiments on aligned samples of membrane proteins. Methods Enzymol. 2005;394:350–382. doi: 10.1016/S0076-6879(05)94014-7. [DOI] [PubMed] [Google Scholar]
- 22.De Angelis AA, Nevzorov AA, Park SH, Howell SC, Mrse AA, Opella SJ. High-resolution NMR spectroscopy of membrane proteins in aligned bicelles. J. Am. Chem. Soc. 2004;126:15340–15341. doi: 10.1021/ja045631y. [DOI] [PubMed] [Google Scholar]
- 23.Mori S, Abeygunawardana C, Johnson MO, Vanzijl PCM. Improved Sensitivity of HSQC Spectra of Exchanging Protons at Short Interscan Delays Using a New Fast HSQC (FHSQC) Detection Scheme That Avoids Water Saturation. J. Magn. Reson. B. 1995;108:94–98. doi: 10.1006/jmrb.1995.1109. [DOI] [PubMed] [Google Scholar]
- 24.Levitt MH, Suter D, Ernst RR. Spin Dynamics and Thermodynamics in Solid-State NMR Cross-Polarization. J. Chem. Phys. 1986;84:4243–4255. [Google Scholar]
- 25.Nevzorov AA, Opella SJ. A “magic sandwich” pulse sequence with reduced offset dependence for high-resolution separated local field spectroscopy. J. Magn. Reson. 2003;164:182–186. doi: 10.1016/s1090-7807(03)00240-4. [DOI] [PubMed] [Google Scholar]
- 26.Fung BM, Khitrin AK, Ermolaev K. An improved broadband decoupling sequence for liquid crystals and solids. J. Magn. Reson. 2000;142:97–101. doi: 10.1006/jmre.1999.1896. [DOI] [PubMed] [Google Scholar]
- 27.Sinha N, Grant CV, Wu CH, De Angelis AA, Howell SC, Opella SJ. SPINAL modulated decoupling in high field double- and triple-resonance solid-state NMR experiments on stationary samples. J. Magn. Reson. 2005 doi: 10.1016/j.jmr.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 28.De Angelis AA, Howell SC, Nevzorov AA, Opella SJ. Structure Determination of a Membrane Protein with Two Trans-membrane Helices in Aligned Phospholipid Bicelles by Solid-State NMR Spectroscopy. J. Am. Chem. Soc. 2006;128:12256–12267. doi: 10.1021/ja063640w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cavanagh J. Protein NMR spectroscopy : principles and practice. Academic Press; San Diego: 1996. [Google Scholar]
- 30.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 31.Goddard TD, Kneller DG. SPARKY 3. University of California; San Francisco: 2004. [Google Scholar]
- 32.Surrey T, Jahnig F. Refolding and oriented insertion of a membrane protein into a lipid bilayer. Proc. Natl. Acad. Sci. U. S. A. 1992;89:7457–7461. doi: 10.1073/pnas.89.16.7457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bayer R, Feigenson GW. Reconstitution of M13 bacteriophage coat protein. A new strategy to analyze configuration of the protein in the membrane. Biochim. Biophys. Acta. 1985;815:369–379. doi: 10.1016/0005-2736(85)90363-3. [DOI] [PubMed] [Google Scholar]
- 34.Marassi FM, Ramamoorthy A, Opella SJ. Complete resolution of the solid-state NMR spectrum of a uniformly 15N-labeled membrane protein in phospholipid bilayers. Proc. Natl. Acad. Sci. U. S. A. 1997;94:8551–8556. doi: 10.1073/pnas.94.16.8551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Franzin CM, Choi J, Zhai D, Reed JC, Marassi FM. Structural studies of apoptosis and ion transport regulatory proteins in membranes. Magn. Reson. Chem. 2004;42:172–179. doi: 10.1002/mrc.1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tian C, Gao PF, Pinto LH, Lamb RA, Cross TA. Initial structural and dynamic characterization of the M2 protein transmembrane and amphipathic helices in lipid bilayers. Protein Sci. 2003;12:2597–2605. doi: 10.1110/ps.03168503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sanders CR, Prosser RS. Bicelles: a model membrane system for all seasons? Structure. 1998;6:1227–1234. doi: 10.1016/s0969-2126(98)00123-3. [DOI] [PubMed] [Google Scholar]
- 38.Prosser RS, Evanics F, Kitevski JL, Al-Abdul-Wahid MS. Current Applications of Bicelles in NMR Studies of Membrane-Associated Amphiphiles and Proteins. Biochemistry. 2006 doi: 10.1021/bi060615u. [DOI] [PubMed] [Google Scholar]
- 39.Park SH, Prytulla S, De Angelis AA, Brown JM, Kiefer H, Opella SJ. High-resolution NMR spectroscopy of a GPCR in aligned bicelles. J. Am. Chem. Soc. 2006;128:7402–7403. doi: 10.1021/ja0606632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baaden M, Sansom MS. OmpT: molecular dynamics simulations of an outer membrane enzyme. Biophys. J. 2004;87:2942–2953. doi: 10.1529/biophysj.104.046987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ramakrishnan M, Qu J, Pocanschi CL, Kleinschmidt JH, Marsh D. Orientation of beta-barrel proteins OmpA and FhuA in lipid membranes. Chain length dependence from infrared dichroism. Biochemistry. 2005;44:3515–3523. doi: 10.1021/bi047603y. [DOI] [PubMed] [Google Scholar]
- 42.Evanics F, Hwang PM, Cheng Y, Kay LE, Prosser RS. Topology of an outer-membrane enzyme: Measuring oxygen and water contacts in solution NMR studies of PagP. J. Am. Chem. Soc. 2006;128:8256–8264. doi: 10.1021/ja0610075. [DOI] [PubMed] [Google Scholar]
- 43.Mouritsen OG, Bloom M. Models of lipid-protein interactions in membranes. Annu. Rev. Biophys. Biomol. Struct. 1993;22:145–171. doi: 10.1146/annurev.bb.22.060193.001045. [DOI] [PubMed] [Google Scholar]
- 44.White SH, Wimley WC. Hydrophobic interactions of peptides with membrane interfaces. Biochim. Biophys. Acta. 1998;1376:339–352. doi: 10.1016/s0304-4157(98)00021-5. [DOI] [PubMed] [Google Scholar]
- 45.Killian JA. Hydrophobic mismatch between proteins and lipids in membranes. Biochim. Biophys. Acta. 1998;1376:401–415. doi: 10.1016/s0304-4157(98)00017-3. [DOI] [PubMed] [Google Scholar]
- 46.Park SH, Opella SJ. Tilt angle of a trans-membrane helix is determined by hydrophobic mismatch. J. Mol. Biol. 2005;350:310–318. doi: 10.1016/j.jmb.2005.05.004. [DOI] [PubMed] [Google Scholar]