Abstract
Neuregulins are a multi-isoform family of growth factors that activate members of the erbB family of receptor tyrosine kinases. The membrane-anchored isoforms contain the receptor-activating ligand in their extracellular domain, a single membrane-spanning region, and a long cytoplasmic tail. To evaluate the potential biological role of the intracellular domain of the membrane-anchored neuregulin isoforms, we used a domain-specific gene disruption approach to produce a mouse line in which only the region of the neuregulin gene encoding almost the entire intracellular domain was disrupted. Consistent with previous reports in which all neuregulin isoforms were disrupted, the resulting homozygous neuregulin mutants died at E10.5 of circulatory failure and displayed defects in neural and cardiac development. To further understand these in vivo observations, we evaluated a similarly truncated neuregulin construct after transient expression in COS-7 cells. This cytoplasmic tail-deleted mutant, unlike wild-type neuregulin isoforms, was resistant to proteolytic release of its extracellular-domain ligand, a process required for erbB receptor activation. Thus, proteolytic processing of the membrane-bound neuregulin isoforms involved in cranial ganglia and heart embryogenesis is likely developmentally regulated and is critically controlled by their intracellular domain. This observation indicates that erbB receptor activation by membrane-bound neuregulins most likely involves a unique temporally and spatially regulated “inside-out” signaling process that is critical for processing and release of the extracellular-domain ligand.
Neuregulins (NRGs) activate erbB receptor protein tyrosine kinases and are involved in neural and heart morphogenesis as well as cell growth and differentiation. They are a diverse multi-isoform protein family encoded by a single gene. Over 15 distinct isoforms have been identified. They result from alternate splicing or from initiation of transcription at different sites (1). The various isoforms give rise to either secreted or membrane-anchored proteins. The latter are composed of an N-terminal extracellular domain, a single transmembrane segment, and a cytoplasmic tail. The secreted isoforms lack transmembrane and cytoplasmic domains. The NRG ligand has been identified as an epidermal growth factor (EGF)-like region (2–5). In the membrane-anchored isoforms, the N-terminal segment of the extracellular domain containing the EGF-like ligand region activates erbB receptors either after proteolytic release or by direct receptor interaction while still attached to its membrane-anchored precursor (6–8). The processes regulating proteolytic release of the extracellular-domain ligand remain unclear but likely involve a signaling event because cleavage can be activated by phorbol esters that stimulate the intracellular effector, protein kinase C (6). Interestingly, the cytoplasmic tails of membrane-anchored NRG isoforms are unusually long (≥150 amino acids). Whether they are involved in mediating bidirectional signaling after binding of the extracellular-domain ligands with their cognate receptors, as is observed for other membrane-anchored growth factors such as Eph, remains to be determined.
To date insights into the biological functions of NRGs have resulted from studies of cultured cell lines or from gross gene-disruption studies in which either erbB receptors or all NRG isoforms have been inactivated. The former provide evidence that NRGs mediate an array of biological effects, including induction of skeletal muscle synthesis of the acetylcholine receptor at the neuromuscular junction (4), stimulation of Schwann cell survival and growth (9), and stimulations of neuronal progenitor cell differentiation (10). Gene-disruption studies, on the other hand, indicate that NRGs are essential for cardiac and cranial ganglia development. Thus, mouse embryos in which both alleles of the NRG gene have been disrupted do not survive past 10.5 days of development (E10.5) and display abnormalities in the development of neural crest cell-derived cranial ganglia and cardiac trabeculae (11, 12). These defects are similar to those observed in erbB2 gene null mutation homozygotes (13) and overlap with those observed with inactivation of both alleles of the erbB4 receptor gene (14).
It is clear from these considerations that in addition to mediating a diverse array of important biological functions, the complexities of NRG structure and biosynthesis present particular challenges for the elucidation of the domain-specific functions and biological roles of the various isoforms of this multi-isoform protein family. Here we report a study in which gene manipulation strategies have been used to address these issues. Specifically, in this paper we have applied a domain-specific gene-disruption approach to inactivate only one domain of a restricted set of NRG isoforms. The results of these studies, coupled with in vitro NRG mutagenesis studies, indicate that the membrane-anchored isoforms are critical for cardiac and neural embryogenesis. Together with the previously reported gene-disruption studies in which either all NRG isoforms or erbB receptors were inactivated, these studies, as a corollary, also provide evidence that the secreted NRG isoforms are most likely not involved in cranial ganglia development or in development of the cardiac trabeculae. Furthermore, we demonstrate that the NRG intracellular domain critically regulates proteolytic release of the extracellular-domain ligand and thus NRG signaling. Again, as a corollary of these findings, it is evident that activation of the erbB receptors regulating cardiac and cranial ganglia development cannot result from a direct interaction between membrane-anchored ligands and receptors but must involve initial proteolytic release of the NRG extracellular-domain ligand.
MATERIALS AND METHODS
Targeting Vector and Intracellular-Domain Deletion Vector Construction.
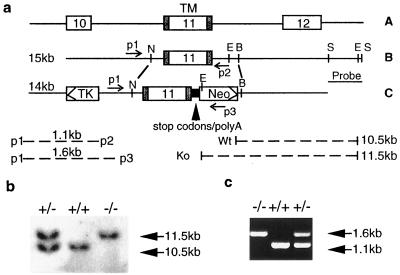
By using a 762-bp BamHI–HindIII fragment of the rat NDF-α2c cDNA to screen a 129/Sv mouse genomic DNA library (Stratagene), a 15-kb genomic clone including exon 11, which encodes the transmembrane domain of the NRG gene, was isolated (Fig. 1a). To specifically disrupt the intracellular domain of NRGs, a 3-kb NheI–BamHI fragment of the 15-kb clone that includes exon 11 was manipulated in vitro by inserting a synthesized DNA fragment immediately 3′ to the coding region for the first three amino acids of the intracellular domain. The fragment contains three stop codons for the three reading frames and a poly(A) sequence from the vector, pcDNA3 (Invitrogen). A neomycin (neo)-resistance gene (15) was inserted downstream of the poly(A) sequence by replacing the partial intron sequence after exon 11. A thymidine kinase expression cassette (16) was also fused 5′ to the intronic sequence upstream of exon 11. A 3′ ScaI–ScaI fragment in the 15-kb clone was deleted from the construct and used as a probe for Southern blot analysis. An intracellular-domain deletion neu differentiation factor (NDF)-construct (ΔCT) was developed by using PCR with primers corresponding to the N terminus and intracellular domain of NDF. The PCR product was inserted into pcDNA3 vector (Invitrogen). The truncated NDF, like the domain-deleted NRG gene construct used for in vivo gene targeting (see above), encodes only the first 3 of the 150 residues of the intracellular domain.
Figure 1.
Disruption of the cytoplasmic tail (CT) of NRGs by gene targeting. (a) Diagram of the partial neuregulin genomic structure (A). A mouse genomic 15-kb clone (B) with exon 11, which encodes the transmembrane domain (TM) was used to generate the targeting construct (C). Restriction enzyme sites: E, EcoRI; B, BamHI; S, ScaI; and N, NheI. Homologous recombination results in acquisition of the EcoRI site from the neo-resistance gene sequence as a result of replacement of the endogenous EcoRI site in the intron sequence. An NRG probe was used for Southern blot screening of genomic DNA digested with EcoRI. The expected size of PCR products (bottom left) generated with the various primers (p1, p2, and p3, see text) is shown. The size of the wild-type and targeted DNA fragments between the EcoRI sites are shown at the bottom right of a. (b) An 11.5kb-fragment was generated from the mutated allele and a 10.5-kb fragment was from the wild type allele. (c) Genotype analysis of E10.5 offspring was performed by PCR using oligonucleotides p1, p2, and p3 and yolk sac DNA. A 1.6-kb fragment was generated from the mutated allele and a 1.1-kb fragment was from the wild-type allele.
Generation and Genotype Analysis of the NRG Mutant Mouse.
The linearized targeting vector was transfected into CJ1 embryonic stem cells, and G418-resistant embryonic stem-cell clones were screened by Southern blot analysis. Recombinant clones were identified and injected into C57BL/6 mouse blastocysts to create chimeric animals. Chimerae were mated with C57BL/6 animals for germ-line transmission of the mutant allele to obtain F1 heterozygotes, which were screened by analyzing tail DNA using Southern blot analysis and PCR. Mating of the heterozygous animals with C57BL/6 mice was continued to increase the number of animals with a mixed-strain background (contributed from 129/Sv and C57BL/6). Embryos resulting from the mating of heterozygotes were analyzed by Southern blotting or PCR using embryo yolk sac DNA as template. Three primers were used for PCR as follows (Fig. 1a): one 5′ end primer from the intron sequence (p1, 5′-AAC AGC CTG ACT GTT AAC ACC) and two 3′ end primers: 5′-CCT GCC TAA GAT GCT TCT ACG TT (p2, from the intron sequence) and 5′-TGC TGT CCA TCT GCA CAG GAC TA (p3, from the neo-resistance gene sequence). DNA samples for Southern blot analysis and PCR were prepared by using standard procedures (17).
Expression of Mutant NRG mRNA.
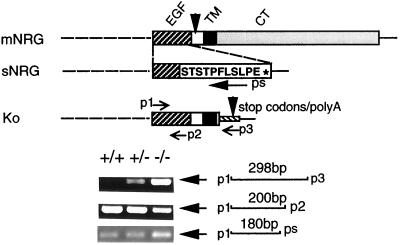
RNA samples were isolated from E10.5 embryos by using a single-step method. Single-stranded DNA was synthesized by using random primers, and PCR was performed according to the manual of the reverse transcription–PCR (RT-PCR) kit (Gibco/BRL). Three specific primers were designed (as shown in Fig. 2): one 5′ end primer based on the N-terminal sequence of the EGF-like domain: p1, 5′-ATA AAG TGT GCG GAG AAG GAG AA; two 3′ end primers: p2, 5′-CCT CTT CTG GTA GAG TTC CTC based on the 3′-end sequence of the EGF-like domain, p3, 5′-TCA GCG AGC TCT AGC ATT TAG based on the sequence of the mutant DNA, ps, CAG AAA GGG AGT GGA CGT ACT based on the unique 3′ coding region of all secreted NRG isoforms. Primers p1/p2 should give products in both mutant and wild-type mice, primers p1/p3 should give a product only with RNA transcribed from the mutated NRG gene, and primers p1/ps should give products in both mutant and wild-type mice, if the mutation does not interfere with the slicing that generates secreted NRG isoforms.
Figure 2.
Evaluation of wild-type and mutant NRG gene transcription. RT-PCR was performed with the primers p1 (corresponding to the 5′ end of the EGF-like domain) and p3 (corresponding to the mutated gene sequence) to demonstrate the mutant NRG mRNA (298 bp), which is present in both heterozygotes and homozygotes but not in the wild-type embryo. A 200-bp fragment was amplified with p1 and p2 (corresponding to the 3′ end sequence of EGF-like domain) from all genotypes to demonstrate gene integrity. A 180-bp fragment was amplified with p1 and ps (corresponding to the unique 3′ coding region of secreted NRG isoforms) from all genotypes, indicating correct expression of all secreted NRG isoforms. mNRG, membrane-anchored NRG gene; sNRG, secreted NRG gene; Ko, truncated NRG gene in NRG gene-targeted animals; TM, transmembrane domain; CT, cytoplasmic tail.
Histologic Analysis and Photography.
E10.5 embryos were isolated by using a stereomicroscope and fixed in 4% paraformaldehyde for 3 hr at 4°C. The embryos were dehydrated in a series of ethanol solutions, embedded with paraffin, and cut into 3-μm sagittal sections for staining with hematoxylin and eosin (H&E). Both cardiac and central nervous system structures were viewed under a microscope and photographed.
Whole-Mount Immunochemistry.
Embryos were fixed in methanol/dimethyl sulfoxide (DMSO) (4:1) overnight at 4°C, treated with methanol/DMSO/30% H2O2 (4:1:1) at room temperature for 5 hr, and rehydrated in phosphate-buffered Tween (PBT, PBS plus 0.5% Tween 20) for 30 min. The rehydrated embryos were incubated with anti-NF160 (anti-neurofilament 160, Sigma) monoclonal antibody (1:60) at 4°C overnight in 5% milk/PBT buffer and washed with PBT five times at room temperature for 60 min each time. They were then incubated with anti-mouse IgG-horseradish peroxidase overnight at 4°C, washed five times with PBT, and developed with diaminobenzidine. Embryos were viewed with a stereomicroscope and photographed.
Cell Culture and Western Blot Analysis.
COS-7 cells (2 × 106 in each 10-cm dish) were transfected with various NDF constructs (20 μg of plasmid DNA) by using a Gene Pulser system (Bio-Rad) and cultured overnight in DMEM containing 10% serum. Cells were then washed three times and cultured in serum-free DMEM for 48 hr before harvesting of conditioned medium or preparation of cell lysates. Western blotting was performed by using standard techniques (17) applied either to cell lysates or to conditioned medium that was initially filtered through a 0.2-μm pore size sterile filter unit (Costar) and then concentrated as much as 50-fold by centrifugation through a Centriplus 10 (Amicon) concentrator. Protein concentration was determined by using the Bradford method (Bio-Rad) and 30 μg of protein was loaded into each well. Monoclonal antibody against the extracellular domain of NDF (1:200, Amgen) was used as the primary antibody at a final concentration of 2 μg/ml (1:100), and anti-mouse IgG-conjugated with horseradish peroxidase (Amersham) was used as the secondary antibody (1:3000). After SDS/PAGE, proteins were electroblotted onto Immobilon-P (Millipore) and detected by using the enhanced chemiluminescence system (Amersham).
Immunocytochemistry of Transfected Cells.
COS-7 or neural 2a cells were obtained from the American Type Culture Collection and cultured in DMEM or MEM, each containing 10% fetal bovine serum, respectively. NDF-α2a and NDF-α2c cDNA were cloned into the mammalian expression vector pcDNA3 (Invitrogen). An intracellular domain-truncated mutant (ΔCT) was produced by PCR using the following primers: (i) 5′-GGC GAG GAA TTC ATG TCT GAG CGC AAA GAA (5′ end primer with EcoRI site italicized); (ii) 5′-ATC CTC GAG CTA CCT ACC TAG GTG TTT GCA GTA GGC (3′ end primer with XhoI site italicized). The three constructs were separately transfected into COS-7 and neural 2a cells by using a Gene Pulser II system (Bio-Rad), and the cells were then plated and cultured on cover slides in 10-cm dishes at 2 × 105 cells per slide for 24–48 hr. Cells were fixed and permeabilized in methanol at −20°C for 15 min and subjected to immunostaining with a monoclonal antibody against the extracellular domain of NDF (1:200, Amgen) as the primary antibody, and anti-mouse IgG-FITC (fluorescein isothiocyanate) (1:60, Sigma) as the secondary antibody.
p185neu (erbB Receptor) Tyrosine Phosphorylation Assay.
COS-7 cell-conditioned medium was filtered through a 0.2-μm pore-size sterile filter unit (Costar) and concentrated. The concentrated medium was added to individual wells of a 6-well plate containing 2 × 105 MCF-7 human breast cancer cells (American Type Culture Collection) in each well. After a 10-min incubation at 37°C, the cells were lysed with 1% Nonidet P-40 lysis buffer that contained 10 mM Tris⋅HCl (pH 7.4)/100 mM NaCl/40 mM NaF/2 mM Na3VO4, and the protease inhibitors phenylmethylsulfonyl fluoride (PMSF) (1 mM), aprotinin (10 μg/ml), and leupeptin (10 μg/ml). The cell lysates were subjected to enhanced-chemiluminescence Western blot analysis with anti-phosphotyrosine antibody RC-20 (1:1,000) (Transduction Laboratories, Lexington, KY).
RESULTS
By using gene targeting, a mouse line was developed in which almost all of the intracellular domain of membrane-anchored NRG isoforms were specifically deleted (see Fig. 1a for a diagram of the targeting construct). Heterozygous mice (NRG/CT+/−, the “CT” is used here to refer to the cytoplasmic tail of NRG) in a mixed-strain background (129/Sv and C57BL/6) were identified by Southern blot analysis (Fig. 1b) and by PCR (Fig. 1c). These animals are phenotypically normal and showed no defects in reproduction or fertility. Among 43 offspring obtained by cross-breeding of heterozygotes, 15 had an NRG/CT+/+ genotype and 28 an NRG/CT+/− genotype, giving a ratio of about 1:2. No live homozygotes (NRG/CT−/−) were obtained. However, homozygous animals were detected at E10.5 or earlier stage embryos, indicating that NRG intracellular-domain deletion resulted in embryonic lethality after E10.5. When the hearts from NRG/CT−/− animals were examined grossly, in many instances it was apparent that they were beating slowly and irregularly. To examine whether the truncated NRG gene was still expressed in homozygotes, RT-PCR was performed by using primers specific for the EGF domain (p1) and for an inserted sequence between the stop codons and the Poly(A) sequence (p3) (Fig. 2). An RT-PCR product of the expected size (298 bp) for the mutant transcript was obtained by using total RNA from NRG/CT+/− or NRG/CT−/− embryos but not from wild-type animals. This product was further confirmed by sequencing analysis, thus indicating that transcription of the mutant gene is not interrupted by insertion of the exogenous DNA. A second concern in the targeted embryos was whether splicing of the transcripts for the secreted NRG isoforms that terminate upstream of the transmembrane domain was impaired. A primer (ps) specific for the 3′ coding region of all secreted isoforms was thus used in RT-PCRs to examine expression of secreted isoforms. As shown in Fig. 2, an expected 180-bp PCR product was obtained and confirmed by sequencing analysis using total RNA from wild-type or homozygous embryos. This product indicates that correct splicing of all secreted isoforms was present. As a control for the RT-PCRs, a third set of primers amplifying the EGF-like domain was used successfully to detect both the wild-type and mutant gene transcripts (Fig. 2, p1-p2 for the 200-bp fragment).
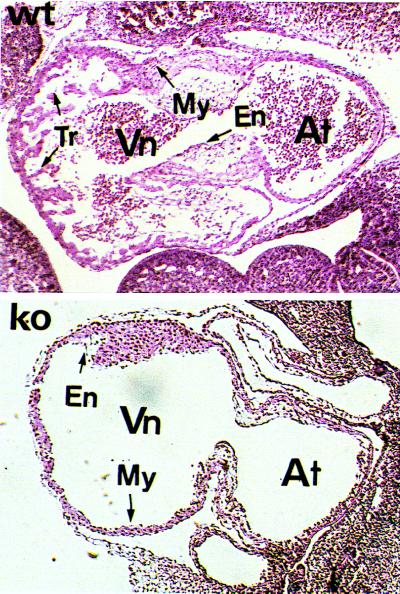
Live embryos at E10.5 were genotyped by PCR, sectioned, and stained with H&E for phenotypic analysis. Compared with the normal hearts, maldevelopment of ventricular trabeculation was evident in E10.5 homozygous embryos (Fig. 3). This defect is consistent with that observed in homozygous embryos (NRG−/−) of a previous NRG-null mutation mouse line, in which the DNA region encoding the receptor-binding domain (the EGF-like domain) of the NRG gene was disrupted (11). Unlike the NRG−/− animals, endocardial cushion development appeared histologically normal in the NRG/CT−/− animals. The reason for this difference is unclear, but it should be noted that involvement of NRGs in endocardial cushion development is contentious, because in contrast with the NRG−/− animals (11), another NRG knock-out mouse line has also been reported not to show endocardial cushion defects (12).
Figure 3.
Abnormal heart development in the mutant mouse. Sagittal sections of H&E-stained heart of the wild-type (upper) and mutant (lower) E10.5 embryos. Vn, ventricle; At, atrium; My, myocardium; En, endocardium; Tr, trabecula.
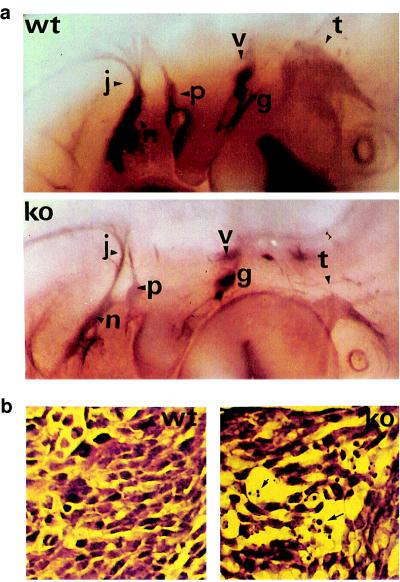
Because NRGs are also expressed in the hindbrain (rhombomers 2, 4, and 6) and the previously reported NRG−/− embryos displayed defects in cranial ganglia development (11), we used whole-mount immunohistochemistry to examine the nervous system. By using anti-NF160, abnormalities of cranial ganglia morphology were observed in E10.5 NRG/CT−/− embryos (Fig. 4a). The dorsal part of the trigeminal ganglion, derived from neural-crest cells, its mandibular branch, and its projections toward the brain stem were absent. The petrose and nodose ganglia were also reduced in size and their projections were hypoplastic and disorganized. In H&E-stained brain sections, the NRG/CT−/− embryos displayed a smaller trigeminal nerve with poor organization and prominent cell death as compared with the wild-type embryos (Fig. 4b). Basically, these defects are similar to those observed in NRG−/−.
Figure 4.
Abnormal development of cranial ganglia in the mutant mouse. (a) Whole-mount immunohistochemistry using an anti-neurofilament antibody to visualize the defects of the nervous system. Cranial ganglia (t, trigeminus; g, geniculate; v, vestibulocochlear; p, petrosal; n, nodose; and j, jugular) are shown in the mutant embryos E10.5 (lower) compared with wild type (upper). (b) H&E-stained sections showing the structure of the trigeminal ganglion in wild-type (left) compared with ko (right) embryos. The ganglion is hypoplastic and poorly developed, with evidence of cell death indicated by arrows.
Thus, NRG/CT−/+ mice are phenotypically normal, whereas NRG/CT−/− animals, in which steady-state mRNA expression from their mutated alleles is normal but the encoded membrane-anchored NRG isoforms lack a cytoplasmic tail, display developmental defects indistinguishable from animals (NRG−/−) in which the NRG ligands of both the membrane-anchored and secreted isoforms, or their cognate erbB receptors, were disrupted. This indicates that (i) expression from at least one normal NRG allele is sufficient for normal neural and cardiac development, (ii) normal development of cranial ganglia and cardiac trabeculae is most likely dependent on the membrane-anchored rather than secreted NRG isoforms, and (iii) that either the cytoplasmic tail of the membrane-anchored NRG isoforms regulates NRG-mediated erbB receptor activation, or is involved in critical bidirectional signaling, or both.
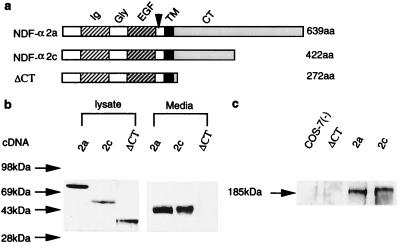
To investigate how the cytoplasmic-tail deletion mutation impairs NRG function, the subcellular localization of NRGs and the proteolytic release of their extracellular-domain ligand were studied in vitro. In these studies, constructs for two wild-type NRG isoforms, NDF-α2a and NDF-α2c, which differ only in the length of their intracellular domain, and for an intracellular domain-truncated mutant, ΔCT (Fig. 5a), were separately transfected into COS-7 Cells. As shown in Fig. 5b, a 43-kDa protein was detected by immunoblot analysis of conditioned culture medium from NDF-α2a- or NDF-α2c-transfected cells, but not from ΔCT-transfected cells. This 43-kDa protein is the proteolytically released, ligand-containing NRG extracellular-domain peptide, because (i) it was detected with an N-terminal-specific NRG antibody, (ii) it is of the predicted Mr for the glycosylated peptide proteolytically released from membrane-anchored NRG isoforms (unpublished data), and (iii) the media from both the NDF-α2a and NDF-α2c but not the ΔCT cells stimulated tyrosine phosphorylation of a 185-kDa erbB receptor protein in MCF-7 human breast cancer cells (Fig. 5c). Importantly, impaired proteolytic release of the extracellular-domain ligand from the ΔCT mutant was not caused by failed expression of the mutant protein, because expression of this protein as well as both wild-type proteins was evident by immunoblot analysis of corresponding COS-7 cell lysates (Fig. 5b).
Figure 5.
In vitro evaluation of NRG extracellular-domain cleavage. Diagram of NDF-α2a, NDF-α2c, and its intracellular domain-truncated mutant ΔCT. (a) Western blot analysis of cell lysate and conditioned media from cells transfected with the indicated cDNA. (b) Stimulation of p185neu tyrosine phosphorylation by the conditioned media from cells transfected with the indicated cDNA or from untransfected cells. (c) Migration position of prestained molecular weight protein standards is shown on the left.
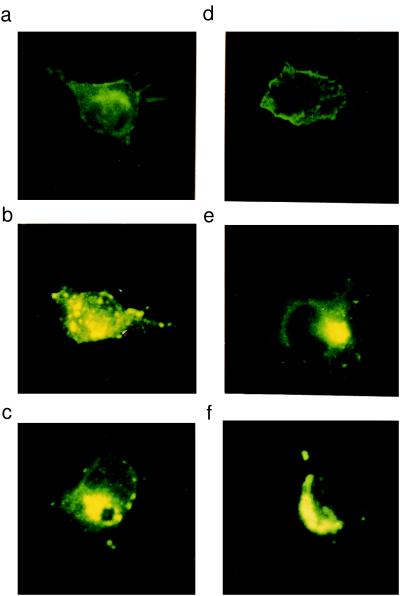
To examine the subcellular localization of the proteins expressed by the ΔCT and wild-type constructs, transfected COS-7 cells were analyzed immunocytochemically. The NDF-α2a-expressed protein localized to the cell membrane and to some extent to the endoplasmic reticulum and Golgi compartments (Fig. 6a). By contrast, the NDF-α2c- and ΔCT-expressed proteins localized predominantly to intracellular organelles (endoplasmic reticulum, Golgi apparati, etc.), with little expression in the cell membrane (Fig. 6 b and c). These results are consistent with previously reported studies of NDF-α2c and NDF-β4a, the latter containing an identical intracellular domain to the NDF-α2a isoform (6). Similar results were obtained with transfection of each of the constructs in the neural 2a cell line (Fig. 6 d–f). Despite the predominant cell-membrane expression of the NDF-α2a isoform and lack of cell-membrane expression of the NDF-α2c isoform, it has been demonstrated previously that both proteins are efficiently processed as a result of intracellular cleavage (18) after transfection in COS-7 cells. The NDF-α2a isoform differs from the NDF-α2c only in that it contains an extra 150 residues at its C terminus; this indicates, along with the 150 residues forming the cytoplasmic tail of the NDF-α2c isoform, that the membrane-proximal C-terminal segment common to both isoforms (but lacking in the ΔCT mutant) is critical for proteolytic release of the extracellular-domain ligand. By contrast, the membrane-distal segment present in the NDF-α2a but not the NDF-α2c isoform (or ΔCT mutant) is required for localization of the former to the cell membrane.
Figure 6.
Cellular localization of NRGs in COS-7 cells and neural 2a cells. COS-7 cells were transfected with NDF-α2a, NDF-α2c, or the intracellular domain truncated mutant, ΔCT and stained with an anti-extracellular domain antibody (a, b, c). NDF-α2a (a) shows strong staining on the cell surface and also some intracellular staining. Cells transfected with NDF-α2c (b) and ΔCT (c) show mainly intracellular staining. The same constructs, 2a (d), 2c (e), and ΔCT (f), were used to transfect neural 2a cells, and similar patterns of expression were observed. In additional studies, immunostaining was performed on transfected cells that were not permeabilized, which confined cell-surface staining only with NDF-α2a-tranfected cells (data not shown).
DISCUSSION
Although previous NRG and erbB receptor-inactivation studies clearly implicated NRG-mediated erbB receptor activation in neural and cardiac embryogenesis, they failed to define the precise NRG domains and specific NRG isoforms involved, because by the nature of the constructs used, erbB receptor signaling by all NRG isoforms was disrupted (11).
In the present study we have used a domain-specific targeting approach to begin to dissect both the domain-specific and isoform-specific contributions to NRG signaling. Because the phenotype of the resulting NRG/CT−/− mice mimic those reported previously with nonselective disruption of all NRG ligands or their cognate receptors, several conclusions can be drawn. First, only the membrane-anchored subset of NRGs and not the secreted isoforms are required for normal cranial ganglia and cardiac trabeculae development. Second, the cytoplasmic tail of the membrane-anchored NRG isoforms is critical for normal NRG signaling. This could be caused by several possibilities, including (i) a requirement of the cytoplasmic tail for adoption of a permissive receptor-active conformation by the NRG extracellular-domain ligand, (ii) regulation of proteolytic processing of the NRG extracellular domain by the cytoplasmic tail, (iii) involvement of the cytoplasmic tail in bidirectional signaling, or (iv) involvement of the cytoplasmic tail in receptor clustering that is essential for some growth factor ligands, such as Eph (19), to mediate cell-to-cell contacts by directly binding to their cognate receptors.
To further understand the role of the cytoplasmic tail of NRGs in signaling and, thus, in neural and cardiac embryogenesis, proteolytic release of the extracellular domain was evaluated after expression of two wild-type NRG constructs (NDF-α2a and NDF-α2c) and a cytoplasmic tail-deleted construct (ΔCT) separately in heterologous, COS-7, or neural 2a cells. In both cultured cell systems proteolytic release of the expected 43-kDa N-terminal ligand-containing peptide was apparent for the wild-type constructs. Furthermore, the released 43-kDa peptide from either construct stimulated tyrosine phosphorylation of 185-kDa erbB receptors in MCF-7 human breast cancer cells. However, neither release of a 43-kDa peptide nor stimulation of tyrosine phosphorylation was evident with the ΔCT construct, despite expression of the encoded, truncated NRG protein.
Resistance of the ΔCT construct to proteolytic processing was not a result of cell membrane expression because both of the wild-type constructs were substrates for proteolytic cleavage, even though immunocytochemical studies clearly indicated predominately cell-membrane expression of the NDF-β4a, but not the NDF-α2c construct (6).
Taken together, these in vitro studies indicate that the juxtamembrane portion of the cytoplasmic tail of membrane-anchored NRG isoforms, which is common to both the NDF-α2a and NDF-α2c isoforms but lacking in the ΔCT construct, critically regulates proteolytic release of the extracellular-domain ligand and thus, signaling by the membrane-anchored isoforms. By contrast, the membrane-distal segment of some membrane-anchored isoforms (such as NDF-α2a) that is lacking in the NDF-α2c isoform, is critical for cellular localization but not for proteolytic release of the extracellular-domain ligand. Furthermore, although deletion of the cytoplasmic tail of membrane-anchored NRG isoforms in the NRG/CT−/− mice likely prevents cell-membrane expression of some NRG isoforms (such as NDF-α2a), this defect in cellular localization alone cannot account for the developmental defects observed in these animals because proteolytic release of the extracellular-domain ligand is intact with isoforms (such as NDF-α2c) that are poorly expressed in the cell membrane. This phenomenon implies that resistance to proteolytic cleavage is sufficient to impair the NRG signaling events required for neural and cardiac development. As a corollary, the cell membrane-expressed NRG isoforms and the potential membrane clustering of these isoforms caused by direct interaction with erbB receptors are not sufficient for the development of these organ systems. This latter contention is supported by the observation that in the developing heart, the endocardium (which expresses NRGs) is widely separated from the myocardium (which expresses erbB receptors), which would thus preclude direct interactions between the membrane-expressed ligands and receptors.
This evidence against a requirement for cell-membrane expression of membrane-anchored NRG isoforms and direct membrane-expressed ligand-receptor interactions also suggests that although the possibility that the cytoplasmic tail may mediate bidirectional signaling by a subset of NRG isoforms cannot be excluded, lack of these isoforms and thus of bidirectional signaling cannot account for the observed developmental defects in NRG/CT−/− mice.
Finally, in contrast with the findings here for the membrane-anchored NRG isoforms, cleavage of the growth factor pro-TGF-α is restricted to the cell membrane. Nevertheless, extracellular-domain processing of pro-TGF-α, like that of the membrane-bound isoforms of NRGs, is regulated by its cytoplasmic tail. With pro-TGF-α, a C-terminal valine residue has been found to be critical for extracellular-domain processing (20). This contrasts with NRGs in which the C-terminal residue is not involved. Rather, involvement of the NRG intracellular domains in protein–protein interactions is required for extracellular-domain processing (21).
In summary, the present studies confirm and extend previous observations implicating NRGs and NRG-mediated erbB receptor activation in cranial-nerve and cardiac development. Specifically, only membrane-anchored and not secreted NRG isoforms appear to be involved in these developmental processes. Furthermore, evidence is presented that these developmental processes are independent of cell-membrane expression, direct membrane-anchored ligand-receptor interactions, and bidirectional signaling, but are critically dependent on intracellular domain-regulated proteolytic release of the NRG extracellular-domain ligand, which is required for NRG signaling.
Acknowledgments
We thank Professor Richard Harvey (Victor Chang Cardiac Research Institute, Sydney, Australia) for critical scientific discussions. We also thank Ms. Elaine Martin and Mrs. Veronica Hammond for expert secretarial assistance. This work was supported in part by Grant 970981 (M.Z. and R.M.G.) from the National Health and Medical Research Council, Australia, and Grant G96S4580 (M.Z. and R.M.G.) from the National Heart Foundation, Australia.
ABBREVIATIONS
- NRG
neuregulin
- EGF
epidermal growth factor
- neo
neomycin
- NDF
neu differentiation factor
- RT-PCR
reverse transcription–PCR
- E
embryonic day
- H&E
hematoxylin and eosin
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1. Peles E, Yarden Y. BioEssays. 1993;15:815–824. doi: 10.1002/bies.950151207. [DOI] [PubMed] [Google Scholar]
- 2.Marchionni M A, Goodearl A D, Chen M S, Bermingham-McDonogh O, Kirk C, Hendricks M, Danehy F, Misumi D, Sudhalter J, et al. Nature (London) 1993;362:312–318. doi: 10.1038/362312a0. [DOI] [PubMed] [Google Scholar]
- 3.Ho W H, Armanini M P, Nuijens A, Phillips H S, Osheroff P L. J Biol Chem. 1995;270:14523–14532. doi: 10.1074/jbc.270.24.14523. [DOI] [PubMed] [Google Scholar]
- 4.Falls D L, Rosen K M, Corfas G, Lane W S, Fischbach G D. Cell. 1993;72:801–815. doi: 10.1016/0092-8674(93)90407-h. [DOI] [PubMed] [Google Scholar]
- 5.Wen D, Peles E, Cupples R, Suggs S V, Bacus S S, Luo Y, Trail G, Hu S, Silbiger S M, Levy R B, et al. Cell. 1992;69:559–572. doi: 10.1016/0092-8674(92)90456-m. [DOI] [PubMed] [Google Scholar]
- 6.Burgess T L, Ross S L, Qian Y X, Brankow D, Hu S. J Biol Chem. 1995;270:19188–19196. doi: 10.1074/jbc.270.32.19188. [DOI] [PubMed] [Google Scholar]
- 7.Morrissey T K, Levi A D, Nuijens A, Sliwkowski M X, Bunge R P. Proc Natl Acad Sci USA. 1995;92:1431–1435. doi: 10.1073/pnas.92.5.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vartanian T, Goodearl A, Viehover A, Fischbach G. J Cell Biol. 1997;137:211–220. doi: 10.1083/jcb.137.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dong Z, Brennan A, Liu N, Yarden Y, Lefkowitz G, Mirsky R, Jessen K R. Neuron. 1995;15:585–596. doi: 10.1016/0896-6273(95)90147-7. [DOI] [PubMed] [Google Scholar]
- 10.Shah N M, Marchionni M A, Isaacs I, Stroobant P, Anderson D J. Cell. 1994;77:349–360. doi: 10.1016/0092-8674(94)90150-3. [DOI] [PubMed] [Google Scholar]
- 11.Meyer D, Birchmeier C. Nature (London) 1995;378:386–390. doi: 10.1038/378386a0. [DOI] [PubMed] [Google Scholar]
- 12.Kramer R, Bucay N, Kane D J, Martin L E, Tarpley J E, Theill L E. Proc Natl Acad Sci USA. 1996;93:4833–4838. doi: 10.1073/pnas.93.10.4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee K F, Simon H, Chen H, Bates B, Hung M C, Hauser C. Nature (London) 1995;378:394–398. doi: 10.1038/378394a0. [DOI] [PubMed] [Google Scholar]
- 14.Gassmann M, Casagranda F, Orioli D, Simon H, Lai C, Klein R, Lemke G. Nature (London) 1995;378:390–394. doi: 10.1038/378390a0. [DOI] [PubMed] [Google Scholar]
- 15.Soriano P, Montgomery C, Geske R, Bradley A. Cell. 1991;64:693–702. doi: 10.1016/0092-8674(91)90499-o. [DOI] [PubMed] [Google Scholar]
- 16.Mansour S L, Thomas K R, Capecchi M R. Nature (London) 1988;336:348–352. doi: 10.1038/336348a0. [DOI] [PubMed] [Google Scholar]
- 17.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. pp. 16.41–16.44. [Google Scholar]
- 18.Meng S-Y, Lu H S, Hu S, Chang D, Yang W, Yanigahara D, Koski R A, Yarden Y. Mol Cell Biol. 1994;14(3):1909–1919. [Google Scholar]
- 19.Bruckner K, Pasquale E B, Klein R. Science. 1977;275:1640–1643. doi: 10.1126/science.275.5306.1640. [DOI] [PubMed] [Google Scholar]
- 20.Bosenberg M W, Pandiella A, Massague J. Cell. 1992;71:1157–1165. doi: 10.1016/s0092-8674(05)80064-9. [DOI] [PubMed] [Google Scholar]
- 21.Liu, X., Hwang, H., Cao, L., Wen, D., Liu, N., Graham, R. M. & Zhou, M. (1998) J. Biol. Chem., in press. [DOI] [PubMed]