Abstract
To study the molecular basis for the clinical phenotype of incomplete penetrance of familial retinoblastoma, we have examined the functional properties of three RB mutations identified in the germ line of five different families with low penetrance. RB mutants isolated from common adult cancers and from classic familial retinoblastoma (designated as classic RB mutations) are unstable and generally do not localize to the nucleus, do not undergo cyclin-dependent kinase (cdk)-mediated hyperphosphorylation, show absent protein “pocket” binding activity, and do not suppress colony growth of RB(−) cells. In contrast, two low-penetrant alleles (661W and “deletion of codon 480”) retained the ability to localize to the nucleus, showed normal cdk-mediated hyperphosphorylation in vivo, exhibited a binding pattern to simian virus 40 large T antigen using a quantitative yeast two-hybrid assay that was intermediate between classic mutants (null) and wild-type RB, and had absent E2F1 binding in vitro. A third, low-penetrant allele, “deletion of RB exon 4,” showed minimal hyperphosphorylation in vivo but demonstrated detectable E2F1 binding in vitro. In addition, each low-penetrant RB mutant retained the ability to suppress colony growth of RB(−) tumor cells. These findings suggest two categories of mutant, low-penetrant RB alleles. Class 1 alleles correspond to promoter mutations, which are believed to result in reduced or deregulated levels of wild-type RB protein, whereas class 2 alleles result in mutant proteins that retain partial activity. Characterization of the different subtypes of class 2 low-penetrant genes may help to define more precisely functional domains within the RB product required for tumor suppression.
To study the molecular basis for the clinical phenotype of incomplete penetrance, we examined the functional properties of a series of retinoblastoma gene (RB) mutations identified in different families with incomplete penetrance. In contrast to classic familial retinoblastoma, which is characterized by a high penetrance for multifocal (bilateral) retinoblastoma tumors, certain kindreds have been reported where germ-line carriers of mutant RB alleles are either clinically unaffected or present with nonproliferative retinal scars (retinomas) or unifocal retinoblastoma (1). The “gatekeeper” gene for retinoblastoma tumors is the RB gene, which encodes a protein product (RB) that plays an important, though yet undefined, role in regulating the transit of dividing cells through the G1/S boundary of the cell cycle and in coordinating cellular differentiation via programmed cell death (apoptotic) pathways in selected tissues. The importance of the link between RB and the cell cycle is highlighted by the following observations: (i) RB is phosphorylated by members of the cyclin-dependent kinase (cdk) system, an enzyme family responsible for driving mitosis in all eukaryotic cells, (ii) cdk-mediated phosphorylation is tightly synchronized with the cell cycle such that RB undergoes hyperphosphorylation just prior to entry into S phase (DNA synthesis), (iii) RB phosphorylation inhibits a protein “pocket” binding activity resulting in the release of a series of cellular protein partners, including members of the E2F transcription factor family, and (iv) all RB mutants isolated from tumor samples (referred here as “classic” RB mutants) have concordantly lost both “pocket” binding activity and the ability to undergo cdk-mediated phosphorylation (2–8). Although RB mutations usually result in an unstable protein product, approximately 10% of lung cancers contain subtle mutations that do not affect RB protein expression (9). In each of these cases, however, functional assays demonstrated 100% loss of protein “pocket” binding activity, cdk-mediated hyperphosphorylation, and the ability to suppress colony growth of RB(−) cells. These findings confirmed that all stable, mutant products isolated from cancer samples have a null phenotype.
In contrast, we recently showed that an RB mutation carried in the germ line of a family with incomplete penetrance of familial retinoblastoma exhibited several unique functional properties (10). This mutant (designated as 661W or Arg661Trp; arginine-to-tryptophan substitution at codon 661) had defective protein-binding activity but retained the ability to undergo cdk-mediated phosphorylation in vivo and the ability to suppress colony growth of RB(−) tumor cells. Although this finding suggested a molecular basis for the phenotype of incomplete penetrance, its significance was limited by the presence of this type of mutation in only a single family. Two additional families, however, that exhibit the phenotype of incomplete penetrance recently have been identified with a germ-line 661W mutation (11). To prove a link between the clinical phenotype of low penetrance and the presence of “partially inactive” RB alleles, we have now studied the functional properties of two additional RB mutations, a deletion of exon 4 (designated Δex4) and a deletion of codon 480 (designated Δ480), identified in distinct kindreds with incomplete penetrance of familial retinoblastoma. Whereas all RB mutants identified from human cancer samples exhibit a null phenotype, we have now demonstrated that three separate RB mutations, identified in at least five different families with incomplete penetrance, represent a unique type of mutation with partial inactivation.
METHODS
Kindreds.
The detailed clinical pedigrees of the families have been previously reported. In summary, a deletion of RB codon 480 mutation (designated here as Δ480) was detected in the germ line of five adult family members, of whom one was unaffected, three had unifocal retinoblastoma, and one had bilateral retinoblastoma (11).
A second family was reported with 58 at-risk family members, of whom 18 had genomic DNA available for analysis (12). In this family, 10 of the 18 family members studied were heterozygous for a germ-line deletion of RB exon 4 (designated here as Δex4). Seven of 10 were clinically unaffected, two had unilateral retinoblastoma, and 1 patient was found to have a retinoma on ophthalmological examination. Of the remaining 40 at-risk family members whose genomic DNA was not available for analysis, 2 patients had bilateral retinoblastoma tumors, 6 had unilateral retinoblastoma, and 2 obligate carriers were clinically unaffected.
We recently characterized the functional properties of a missense mutation at RB codon 661 (10, 13). Since that report, two other families with the phenotype of incomplete penetrance were identified that carried the same germ-line mutation (designated 661W) (11). The largest of these 3 families had 14 at-risk individuals in 3 generations (13). Thirteen family members were available for genetic analysis of germ-line DNA, and 10 of the 13 family members tested were shown to be heterozygous carriers for the 661W mutant allele. Of these 10 heterozygous carriers, 4 adult family members remain clinically unaffected, 2 were found to have only benign retinomas/retinal scars, 2 had unilateral retinoblastoma, and 2 had bilateral retinoblastoma. The two other families with the same 661W mutation had either seven or five family members at-risk who were available for genetic analysis (11). Six of seven members from the first family were heterozygous and showed one unaffected gene carrier, four subjects with unilateral retinoblastoma, and only one with multifocal retinoblastoma. All five tested members of the last family were shown to be heterozygous carriers of the 661W mutation; two were clinically unaffected, and three had unilateral retinoblastoma.
In Vivo RB Phosphorylation Analysis.
cDNA constructs of the “classic” RB mutant alleles (706F, Δ ex21, and Δ ex22) were isolated from small cell lung cancer samples (14, 15). The RB mutations identified from the families with incomplete penetrance were incorporated into paired sets of oligonucleotides that anchored PCR amplifications. Each of the fragments was subcloned into a eukaryotic expression plasmid (pCI-neo; Promega) and confirmed by nucleotide sequencing. The RB constructs were cotransfected with a series of cyclin constructs (16) into the RB(−) H2009 tumor cell line (17), and protein extracts were subjected to sequential α-RB immunoprecipitation and α-RB immunoblot analysis at 72 hr as previously described (15). Stable RB transfectants (without ectopic cyclins) were expanded in the presence of G418, and protein extracts were subjected to sequential immunoprecipitation and immunoblotting analysis as described above. Subcellular localization was performed by subjecting the RB transfectants to immunohistochemical staining using α-RB as previously published (10).
Yeast Two-Hybrid Binding Analysis.
Pocket RB cDNAs (from codon 378 to 792) were generated by PCR amplification for each of the mutant and wild-type RB expression plasmids and cloned in-frame into the Gal 4 DNA-binding domain plasmid pGBT9 (CLONTECH). The pGBT9-RB plasmids were used to transform yeast strain SFY526 with and without cotransfection of a large T cDNA cloned in-frame into the Gal 4 activation domain plasmid (pTD1, CLONTECH) onto appropriate selective media by a lithium acetate method as described by the manufacturer (Bio 101). The yeast strain SFY526 (MATa, ura3–52, his3–200, ade2–101, lys2–801, trp1–901, leu2–3, 112, canr, gal4–542, gal80–538, URA3::GAL1-lacz) was obtained from CLONTECH. A qualitative colony lift filter assay was used to detect β-galactosidase activity, and a quantitative assay using o-nitrophenyl β-d-galactopyranoside (ONPG; Sigma) was performed. One unit of β-galactosidase activity is defined as the amount that hydrolyzes 1 μmol of ONPG to o-nitrophenol and d-galactose per min per cell: β-galactosidase units = 1,000 × OD420/time (min) × volume × concentration factor × OD600. The quantitative liquid β-galactosidase assay was a modification of that described previously (18) as directed by the manufacturer (CLONTECH).
In Vitro E2F1 Binding Analysis.
Wild-type and mutant RB proteins were in vitro translated in the presence of [35S]methionine and incubated with a recombinant glutathione S-transferase (GST)-E2F1 fusion protein that had been prepared and purified as described (19). Bound proteins were washed, separated by SDS/PAGE, and subjected to autoradiography.
Growth Suppression Analysis.
Three micrograms of wild-type and mutant RB cDNA expression plasmids were used to transfect 1 × 106 SaOS cells, and the number of G418-resistant colonies were counted at 3 weeks after fixation and staining with crystal violet as previously published (20).
RESULTS
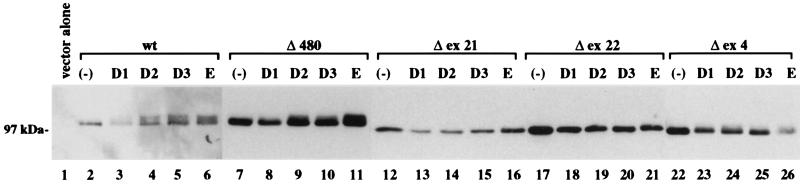
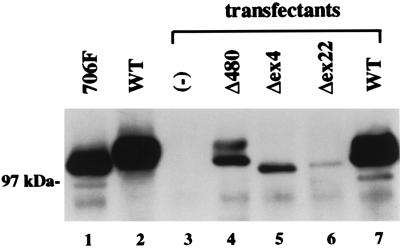
To assess the functional properties of the mutant RB alleles, we performed a series of biochemical and biological assays including an in vivo hyperphosphorylation assay, a nuclear localization assay, a quantitative yeast two-hybrid binding assay with simian virus 40 large T antigen, a protein binding assay with recombinant fusion E2F1, and in vivo colony growth-suppression assay. We observed that the Δ480 RB mutant retained the ability to undergo hyperphosphorylation in vivo in response to the transient coexpression of both ectopic cyclin E or members of the cyclin D family (Fig. 1, lanes 7–11). To examine the ability of the Δ480 RB mutant to undergo hyperphosphorylation under more physiological conditions, we tested cell lysates of stable transfectants growing in mass culture without the coexpression of ectopic cyclins. We observed that the Δ480 RB mutant exhibited a hyperphosphorylation pattern that was indistinguishable from wild-type RB (Fig. 2, lane 4). In addition, these findings were identical to that previously observed with the 661W mutation (10), despite the fact that the Δ480 and 661W mutations are located in different regions of the RB pocket referred to, respectively, as domain A and domain B. In contrast, we showed that two RB mutants isolated from small cell lung cancer samples were unable to undergo hyperphosphorylation despite the coexpression of cyclins D1, D2, D3, or E (Fig. 1, lanes 12–21). This is consistent with previous reports showing that all RB mutants lack the ability to undergo cdk-mediated hyperphosphorylation. Analysis of the third low-penetrant allele, a Δex4 mutation that is located outside of the RB pocket domain near the N terminus of the protein, showed a different pattern with a minimal phosphorylation shift in response to cyclin D2 and cyclin E expression (Fig. 1, lanes 22–26) and no hyperphosphorylation when stably expressed in the absence of ectopic cyclins (Fig. 2, lane 5). Immunohistochemical analyses of the RB(−) H2009 lung tumor line transfected with either 661W (10), Δ480, or Δex4 showed that the mutant proteins localized to the nucleus (data not shown).
Figure 1.
In vivo phosphorylation after transient cotransfection of RB and cyclins. Sequential α-RB immunoprecipitation and immunoblot analysis of protein lysates from the RB(−) tumor line H2009 after transient transfection with either the parental mammalian expression plasmid (lane 1) or the plasmid containing wild-type (wt) RB (lanes 2–6), Δ480 (lanes 7–11), Δex21 (lanes 12–16), Δex22 (lanes 17–21), or Δex4 RB (lanes 22–26). The H2009 cells were cotransfected with different cyclin constructs as indicated, and protein lysates were harvested at 72 hr for analysis.
Figure 2.
In vivo phosphorylation of stable RB transfectants. Sequential α-RB immunoprecipitation and immunoblot analysis showing lysates containing the 706F mutant and wild-type RB product (lanes 1 and 2). Protein lysates from stable transfectants of the H2009 cell line expressing the parental vector alone (lane 3) or the different RB cDNAs without ectopic cyclins are indicated (lanes 4–7).
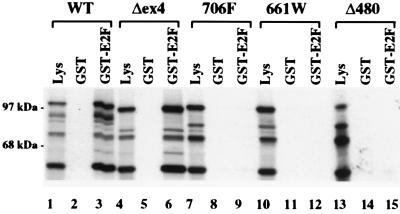
Protein-binding activity, using an in vitro binding assay with recombinant E2F1 as a target, showed that the Δex4 mutant retained substantial E2F1 binding, whereas both the 661W and Δ480 mutants had <5% binding, which was not consistently different than the null binding of the classic mutant controls (Fig. 3). A ladder of protein bands is always observed following in vitro translation of the RB cDNA, where the largest band represents full-length p105 kDa RB and the smaller species represent N-terminal truncations due to either promiscuous translation initiation or proteolysis. As expected, only the two largest species translated from the Δex4 construct are smaller than the corresponding species from wild-type RB mapping the approximate location of the exon 4 deletion (Fig. 3, lane 4).
Figure 3.
In vitro E2F1 binding. [35S]methionine-labeled in vitro-translated RB cDNAs were subjected to SDS/PAGE and autoradiography before (lanes 1, 4, 7, 10, and 13) or after (lanes 2, 5, 8, 11, and 14) precipitation by GST or GST-E2F1 (lanes 3, 6, 9, 12, and 15).
To further examine in vivo binding properties of the 661W and Δ480 mutants we utilized a quantitative yeast two-hybrid assay with a series of different pocket constructs fused to the Gal4 DNA-binding domain. The five-pocket constructs spanned the identical region of domains A and B (codons 378 to 792) and differed only in the single point mutations: Δ480, 661W, 707W (an in vitro-generated serine-to-tryptophan mutant at codon 707, which we previously demonstrated to have slightly reduced E2F1-binding activity in vitro) (21), the null mutant 706F, and wild-type RB. We performed three, independent experiments and demonstrated that the Δ480 and 661W mutants (identified in families with low penetrance) exhibited a binding pattern that was intermediate between 706F (null mutant identified from a lung tumor) and wild-type RB (Table 1). The quantitative bindings of the Δ480 and 661W mutants to large T antigen were 22 and 8.5% of the wild-type RB binding, respectively. In contrast, the in vitro-generated 707W mutant (which is not linked to a clinical sample) showed higher levels of protein binding than the clinically associated low-penetrant alleles.
Table 1.
Yeast two-hybrid assay
| Clone | β-Galactosidase units ± SD
|
|
|---|---|---|
| Alone | With large T-AD | |
| Large T-AD | 4.87 ± 0.90 | |
| p53-BD | 0.45 ± 0.48 | 178.35 ± 11.46 |
| wt RB-BD | 0.59 ± 0.20 | 299.92 ± 19.19 |
| 707W-BD | 0.44 ± 0.39 | 166.72 ± 36.58 |
| Δ480-BD | 0.51 ± 0.10 | 66.75 ± 6.00 |
| 661W-BD | 0.30 ± 0.10 | 25.40 ± 7.76 |
| 706F-BD | 0.46 ± 0.11 | 2.03 ± 0.28 |
One unit of β-galactosidase hydrolyzes 1 μmol of ONPG to o-nitrophenol and d-galactose per min per cell; β-galactosidase units = 1,000 × OD420/time (min) × volume × concentration factor × OD600. AD, Ga14 activation domain plasmid (pGAD); BD, Ga14 DNA-binding domain plasmid (pGBT9).
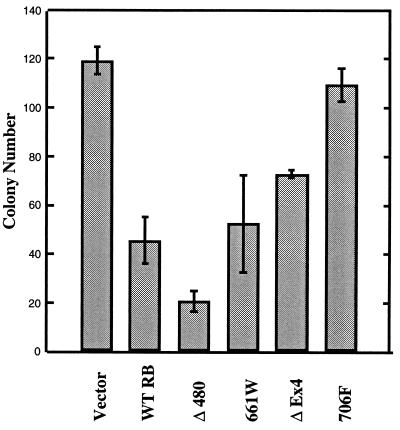
A consistent difference between the low-penetrant mutant RB alleles and all other classic RB mutations could also be detected using an in vivo colony-suppression assay in the SaOS2 osteosarcoma cell line. Although the parental vector alone or the 706F RB mutation (15) showed no growth suppression, we observed colony suppression with wild-type RB and with the incomplete penetrant alleles Δ480, 661W, and Δex4 when expressed in the SaOS2 osteosarcoma cell line (Fig. 4).
Figure 4.
Colony suppression assay. SaOS2 cells were transfected with the parental vector alone (lane 1) or with the different RB cDNAs as indicated (lanes 2–6), and colonies were scored at 3 weeks in the presence of G418 as described in Materials and Methods.
DISCUSSION
Although the RB gene has served as the paradigm for the two-hit theory of cancer, the molecular basis for the clinical phenotype of familial retinoblastoma with incomplete penetrance has been poorly defined. While early hypotheses proposed immunological or other host-mediated factors (22–24), an important clue to a genetic basis for this syndrome was the observation that a germ-line mutation within the promoter region of RB was observed in two different families with low penetrance (25). This report suggested that reduced or deregulated expression of wild-type RB may explain the variable penetrance of tumor formation observed in obligate carriers. In addition, it was proposed that the reduplication of the mutant allele could tip the balance in favor of suppression of the malignant phenotype. Since that initial report, an additional low-penetrant family has been identified that also contains a germ-line mutation within the promoter region of RB (26).
Another approach to the analysis of the phenotype of incomplete penetrance is to directly investigate the functional properties of germ-line RB alleles that contain subtle mutations within the encoded ORF. Although all stable RB mutants from sporadic adult cancers show the concordant loss of both protein binding and hyperphosphorylation, we previously observed that an in vitro generated mutant, 707W, demonstrated the novel phenotype of moderately reduced E2F1 binding with apparently normal cdk-mediated hyperphosphorylation (21). To address whether there might be a “milder” clinical phenotype associated with this dissociation of hyperphosphorylation from protein binding, we examined the functional properties of a series of mutant alleles from different families with the phenotype of incomplete penetrance of familial retinoblastoma. Although we had no assurance that these predicted mutant proteins would be sufficiently stable for functional analyses, we were encouraged by earlier studies with a series of stable “classic” RB mutants isolated from lung cancer samples (9, 14). In each of these tumor cases, however, the abundant steady-state levels of mutant RB protein had no measurable biologic activity and are presumed to be null. Characterization of the functional properties of the three low-penetrant RB alleles also showed stable protein expression, however, with a unique phenotype that retained many wild-type functions including nuclear localization, cdk-mediated phosphorylation, retention of intermediate levels of large T antigen binding, and the ability to suppress colony growth when overexpressed in RB(−) cells. Each of these families was characterized as having low penetrance by the presence of retinomas and by a reduced disease eye ratio (der), which is defined as the ratio of the sum of affected eyes to the sum of mutation carriers (11). The der of these five families ranged from 0.2 to 1.0, whereas classic familial retinoblastoma usually have der scores that approach 2.0 and are at least 1.5.
The association of a specific type of RB mutant allele with the phenotype of incomplete penetrance raises several important points. First, it provides compelling evidence that the phenotype for this clinical presentation is encoded by the specific type of RB gene mutation. These low-penetrant alleles can be classified into two general categories: class 1 alleles, representing genetic [or possibly epigenetic, i.e., methylation (27)] changes within regulatory promoter regions resulting in reduced or deregulated levels of wild-type protein or class 2 alleles, representing a mutant protein product that retains some parameters of tumor-suppressor activity. An important issue suggested by this data is whether there is a single class 2 low-penetrant phenotype or whether there are many different subtypes of class 2 alleles corresponding to the separate domains targeted in different low-penetrant families. Although all three low-penetrant alleles induced colony suppression of an RB(−) tumor line, our data suggested clear differences in biochemical properties between them. For example, the Δ480 (located in pocket domain A) and the 661W (located in domain B) alleles shared similar functional properties, including the ability to undergo hyperphosphorylation in vivo with or without cyclin coexpression and the absence of E2F1 binding in vitro. The Δex4 allele, however, showed only minimal phosphorylation following overexpression of cyclin D2 or cyclin E and no detectable hyperphosphorylation in the absence of ectopic cyclin expression, and it retained E2F1 binding in vitro. These findings further emphasize the dissociation of cdk-mediated phosphorylation from “pocket” protein-binding activity, which characterizes each of the low-penetrant alleles and is never observed in the classic RB mutants. In addition, the observation that the Δex4 protein retains all of the major cdk phosphorylation sites (28, 29) suggests that the functional activity of the constitutively hypophosphorylated Δex4 product may not be analogous to the reversibly hypophosphorylated wild-type RB. Recent studies have suggested a role for the N-terminal domain of RB to bind a family of cellular proteins, including an enzyme with RB and histone H1 kinase activity, which is maximally active during the G2/M phase of the cell cycle (30, 31). Whether the disruption of this N-terminal binding activity underlies the clinical syndrome in patients carrying the Δex4 allele, however, remains to be defined.
The biochemical analyses of the low-penetrant RB alleles suggests two hypotheses to understand the genotype:phenotype relationship for incomplete penetrance of familial retinoblastoma. In the simplest model, a threshold level of a single RB pocket binding activity is solely responsible for tumor suppression and this “pocket threshold” may be jeopardized by “partially active” alleles using complementary mechanisms, including reduced mRNA expression, reduced protein stability, inefficient compartmental targeting, or reduced protein pocket binding affinity. Biochemical analyses of the pocket mutants, 661W and Δ480, are consistent with this interpretation; however, this model would require that the Δex4 product has a defect in pocket activity that is not detectable by our in vitro assays. In addition, this model would incorporate families with incomplete penetrance that result from germ-line mutations within the regulatory regions of the RB promoter (25, 26). An alternative view proposes a complex, multifunctional RB molecule where at least two discrete functions/domains act in concert to mediate tumor suppression. This suggests that loss of E2F1 binding would need to cooperate with the loss of another undefined RB function to result in the null phenotype. This hypothesis predicts that the mutations identified in classic familial retinoblastoma or sporadic adult human cancers have a more global effect on RB functions with concordant loss of both cdk-mediated phosphorylation and protein-binding activity. In contrast, the more subtle low-penetrant RB mutations are predicted to have selectively targeted a single domain that is characterized (at least with the low-penetrant alleles studied in this report) by a discordance between phosphorylation in vivo and pocket binding. Regardless of the underlying mechanism, the additional recruitment of RB activity during a hypothetical window of vulnerability, for example, through mRNA induction, might shift functional activity at the cellular level by superseding these defects. In addition, it is possible that this putative induction of RB activity may underlie the etiology of the benign retinal scars (retinomas), although direct evidence to test this hypothesis will require a suitable animal model. This model of a partially inactivated gatekeeper gene could be of general interest because the phenomenon of rare, spontaneous tumor regressions has been observed in several tumor types besides retinoblastoma. These observations also raise other clinical speculations. The finding that different families carry the same 661W mutation suggests that certain low-penetrant alleles will be the single, most commonly reported RB mutations in retinoblastoma. Future studies will be required to address whether retinal tumors in infants carrying selected low-penetrant alleles would have a less aggressive course, with the retinoma at the extreme end of the spectrum. These findings, therefore, raise the question of whether the response to nonsurgical therapies, such as chemotherapy, radiotherapy, or observation alone, would be greater in patients with low-penetrant alleles than in those with a “classic” RB mutation.
Acknowledgments
We thank Steve Dowdy for the cyclin plasmids, Lisa Pogoda for technical assistance, and Ilan Kirsch, William Sellers, and William Kaelin, Jr., for review of the manuscript.
ABBREVIATIONS
- cdk
cyclin-dependent kinase
- Δ480
deletion of codon 480
- Δex4
deletion of exon 4
- 661W
arginine-to-tryptophan substitution at codon 661
Footnotes
To whom reprint requests should be addressed at: National Cancer Institute–Navy Oncology, Bldg. 8/Rm 5101, Naval Hospital, Bethesda, MD 20889. e-mail: fkaye@helix.nih.gov.
References
- 1.Gallie B L, Ellsworth R M, Abramson D H, Phillips R A. Br J Cancer. 1982;45:513–521. doi: 10.1038/bjc.1982.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buchkovich K, Duffy L A, Harlow E. Cell. 1989;58:1097–1105. doi: 10.1016/0092-8674(89)90508-4. [DOI] [PubMed] [Google Scholar]
- 3.Chen P L, Scully P, Shew J Y, Wang J Y, Lee W H. Cell. 1989;58:1193–1198. doi: 10.1016/0092-8674(89)90517-5. [DOI] [PubMed] [Google Scholar]
- 4.DeCaprio J A, Ludlow J W, Lynch D, Furukawa Y, Griffin J, Piwnica-Worms H, Huang C M, Livingston D M. Cell. 1989;58:1085–1095. doi: 10.1016/0092-8674(89)90507-2. [DOI] [PubMed] [Google Scholar]
- 5.Ludlow J W, DeCaprio J A, Huang C M, Lee W H, Paucha E, Livingston D M. Cell. 1989;56:57–65. doi: 10.1016/0092-8674(89)90983-5. [DOI] [PubMed] [Google Scholar]
- 6.Lin B T, Gruenwald S, Morla A O, Lee W H, Wang J Y. EMBO J. 1991;10:857–864. doi: 10.1002/j.1460-2075.1991.tb08018.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chellappan S P, Hiebert S, Mudryj M, Horowitz J M, Nevins J R. Cell. 1991;65:1053–1061. doi: 10.1016/0092-8674(91)90557-f. [DOI] [PubMed] [Google Scholar]
- 8.Weinberg R A. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 9.Shimizu E, Coxon A, Otterson G A, Steinberg S M, Kratzke R A, Kim Y W, Fedorko J, Oie H, Johnson B E, Mulshine J L, Minna J D, Gazdar A F, Kaye F J. Oncogene. 1994;9:2441–2448. [PubMed] [Google Scholar]
- 10.Kratzke R A, Otterson G A, Hogg A, Coxon A B, Geradts J, Cowell J K, Kaye F J. Oncogene. 1994;9:1321–1326. [PubMed] [Google Scholar]
- 11.Lohmann D R, Brandt B, Hopping W, Passarge E, Horsthemke B. Hum Genet. 1994;94:349–354. doi: 10.1007/BF00201591. [DOI] [PubMed] [Google Scholar]
- 12.Dryja T P, Rapaport J, McGee T L, Nork T M, Schwartz T L. Am J Hum Genet. 1993;52:1122–1128. [PMC free article] [PubMed] [Google Scholar]
- 13.Onadim Z, Hogg A, Baird P N, Cowell J K. Proc Natl Acad Sci USA. 1992;89:6177–6181. doi: 10.1073/pnas.89.13.6177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horowitz J M, Park S H, Bogenmann E, Cheng J C, Yandell D W, Kaye F J, Minna J D, Dryja T P, Weinberg R A. Proc Natl Acad Sci USA. 1990;87:2775–2779. doi: 10.1073/pnas.87.7.2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaye F J, Kratzke R A, Gerster J L, Horowitz J M. Proc Natl Acad Sci USA. 1990;87:6922–6926. doi: 10.1073/pnas.87.17.6922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dowdy S F, Hinds P W, Louie K, Reed S I, Arnold A, Weinberg R A. Cell. 1993;73:499–511. doi: 10.1016/0092-8674(93)90137-f. [DOI] [PubMed] [Google Scholar]
- 17.Kratzke R A, Shimizu E, Geradts J, Gerster J L, Segal S, Otterson G A, Kaye F J. Cell Growth Differ. 1993;4:629–635. [PubMed] [Google Scholar]
- 18.Yocum R R, Hanley S, West R J, Ptashne M. Mol Cell Biol. 1984;4:1985–1998. doi: 10.1128/mcb.4.10.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaelin W, Jr, Pallas D C, DeCaprio J A, Kaye F J, Livingston D M. Cell. 1991;64:521–532. doi: 10.1016/0092-8674(91)90236-r. [DOI] [PubMed] [Google Scholar]
- 20.Qin X Q, Chittenden T, Livingston D M, Kaelin W., Jr Genes Dev. 1992;6:953–964. doi: 10.1101/gad.6.6.953. [DOI] [PubMed] [Google Scholar]
- 21.Kratzke R A, Otterson G A, Lin A Y, Shimizu E, Alexandrova N, Zajac-Kaye M, Horowitz J M, Kaye F J. J Biol Chem. 1992;267:25998–26003. [PubMed] [Google Scholar]
- 22.Gallie B L, Wong J J, Ellsworth R M, Dupont B, Good R A. In: Immunology and Immunopathology of the Eye. Silverstein A, O’Connor G, editors. New York: Masson; 1979. [Google Scholar]
- 23.Char D H, Ellsworth R, Rabson A S, Albert D M, Herberman R B. Am J Ophthalmol. 1974;78:5–11. doi: 10.1016/0002-9394(74)90003-8. [DOI] [PubMed] [Google Scholar]
- 24.Matsunaga E. Am J Hum Genet. 1978;30:406–424. [PMC free article] [PubMed] [Google Scholar]
- 25.Sakai T, Ohtani N, McGee T L, Robbins P D, Dryja T P. Nature (London) 1991;353:83–86. doi: 10.1038/353083a0. [DOI] [PubMed] [Google Scholar]
- 26.Cowell J K, Bia B, Akoulitchev A. Oncogene. 1996;12:431–436. [PubMed] [Google Scholar]
- 27.Greger V, Passarge E, Höpping W, Messmer E, Horsthemke B. Hum Genet. 1989;83:155–158. doi: 10.1007/BF00286709. [DOI] [PubMed] [Google Scholar]
- 28.Lin B T-Y, Gruenwald S, Morla A O, Lee W-H, Wang J Y J. EMBO J. 1991;10:857–864. doi: 10.1002/j.1460-2075.1991.tb08018.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lees J A, Buchkovich K J, Marshak D R, Anderson C W, Harlow E. EMBO J. 1991;10:4279–4290. doi: 10.1002/j.1460-2075.1991.tb05006.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sterner J M, Murata Y, Kim H G, Kennett S B, Templeton D J, Horowitz J M. J Biol Chem. 1995;270:9281–9288. doi: 10.1074/jbc.270.16.9281. [DOI] [PubMed] [Google Scholar]
- 31.Sterner J M, Tao Y, Kennett S B, Kim H G, Horowitz J M. Cell Growth Differ. 1996;7:53–64. [PubMed] [Google Scholar]