Abstract
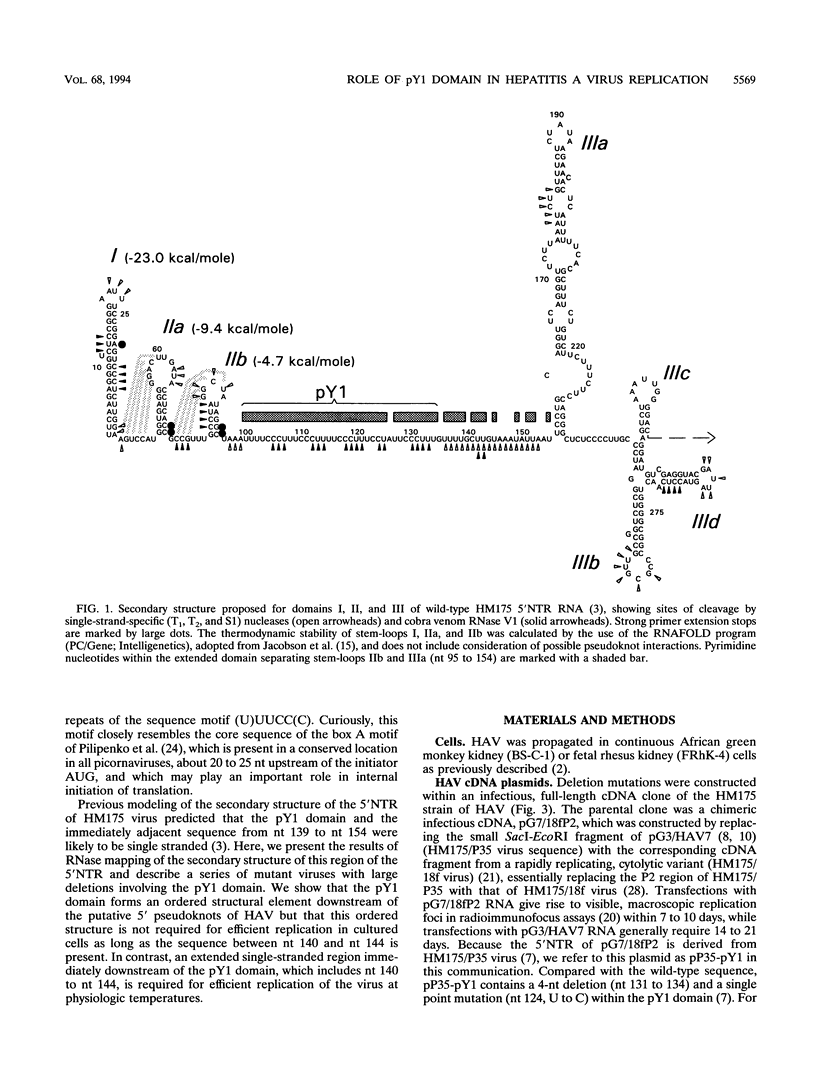
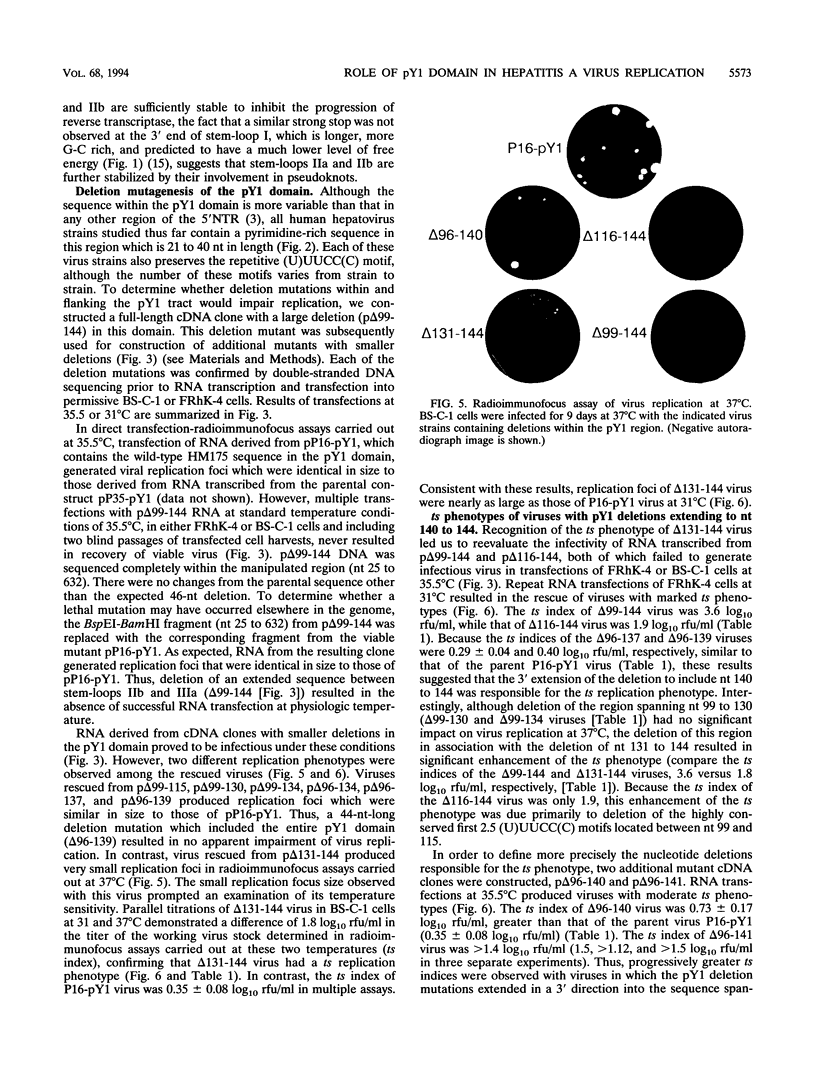
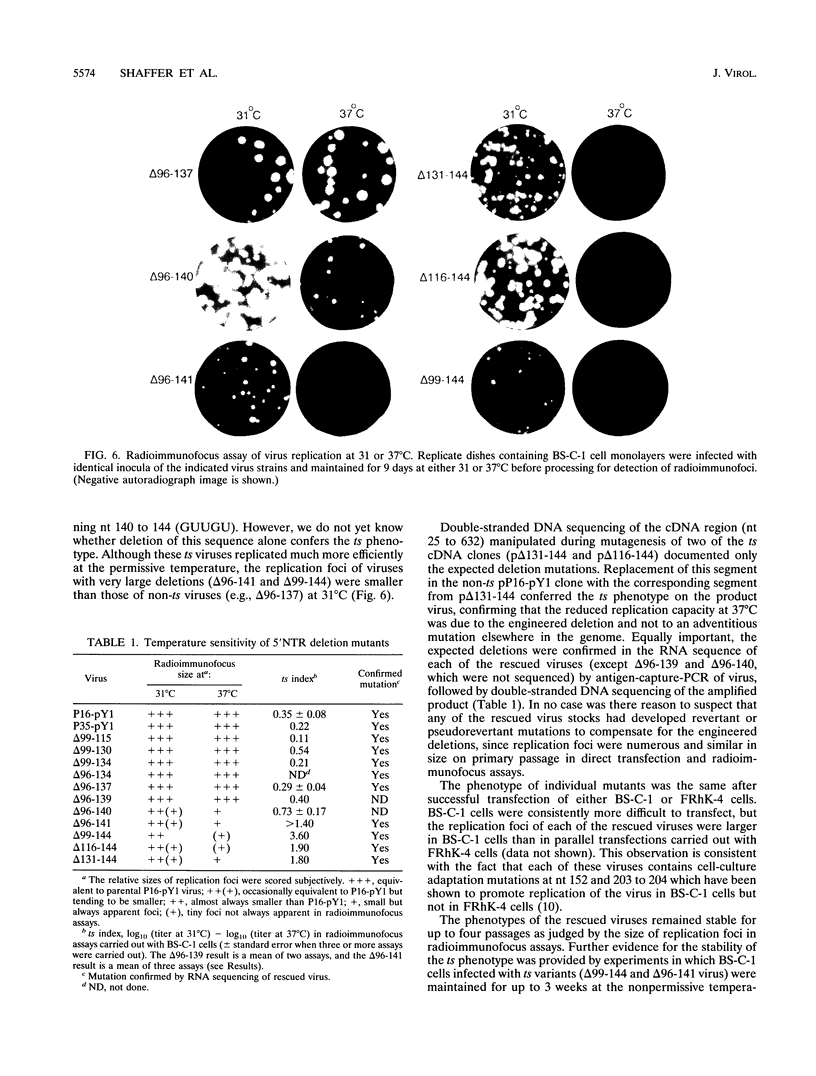
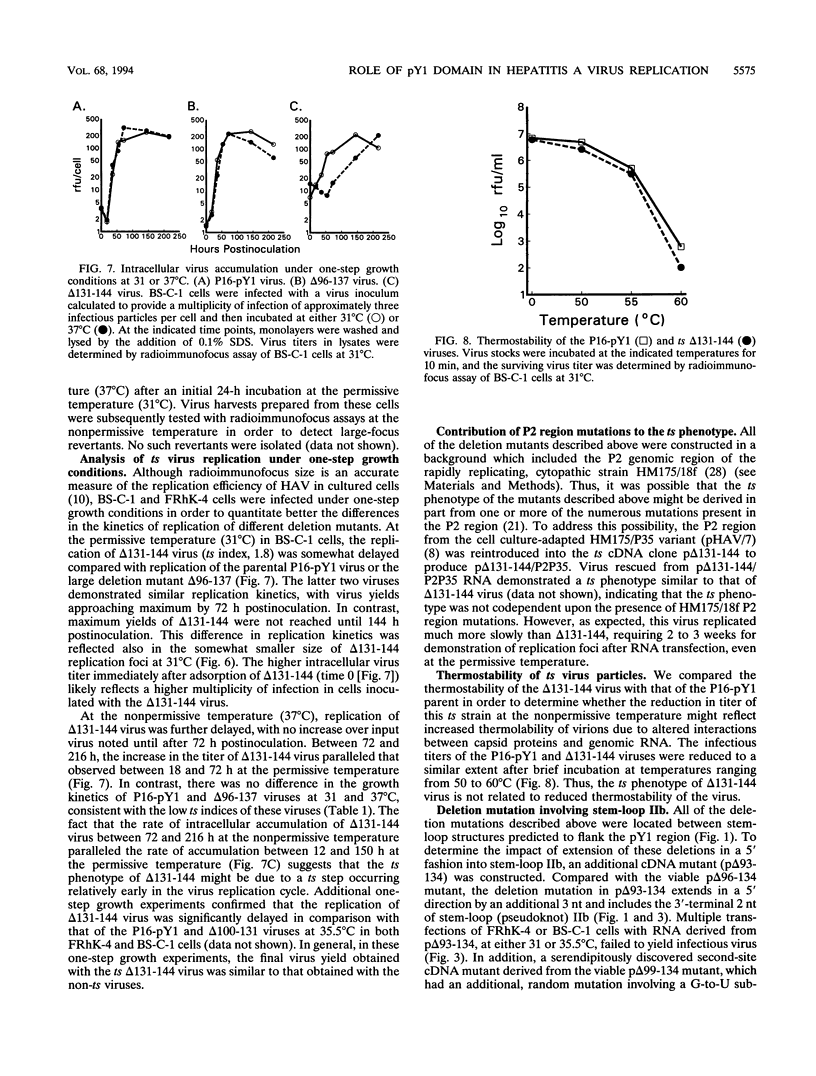
The 5' nontranslated RNA (5'NTR) of the HM175 strain of human hepatitis A virus contains several pyrimidine-rich regions, the largest and most 5' of which (pY1) is an almost pure polypyrimidine tract located between nucleotides (nt) 99 and 138, which includes five tandem repeats of the sequence motif (U)UUCC(C). Previous modeling of the RNA secondary structure suggested that this region was likely to be single-stranded, but repetitive RNase V1 cleavage sites within these (U)UUCC(C) motifs indicated that pY1 possesses an ordered structure. To assess the role of this domain in replication of the virus, a series of large deletion mutations were created which involved the pY1 domain of an infectious cDNA clone. Deletion of 44 nt between nt 96 and 139, including the entire pY1 domain, did not reduce the capacity of the virus to replicate in BS-C-1 or FRhK-4 cells, as assessed by the size of replication foci in radioimmunofocus assays or by virus yields under one-step growth conditions. In contrast, viable virus could not be recovered from transfected RNAs in which the deletion was extended in a 5' direction by an additional 3 nt (delta 93-134), most likely because of the destabilization of a predicted stem-loop structure upstream of pY1. Deletion mutations extending in a 3' fashion to nt 140, 141, or 144 resulted in moderately (delta 96-140 and delta 96-141) or strongly (delta 99-144, delta 116-144, and delta 131-144) temperature-sensitive replication phenotypes. Although deletion of the pY1 domain did not by itself affect the replication phenotype of virus, the additional deletion of sequence elements within the pY1 domain (nt 99 to 130) substantially enhanced the temperature-sensitive phenotype of delta 131-144 virus. These data suggest that the (U)UUCC(C) motifs within the pY1 domain are conserved among wild-type viruses in order to serve a function required during infection in vivo but not in cell culture. In contrast, the single-stranded region located immediately downstream of pY1 (nt 140 to 144) is essential for efficient replication in cultured cells at physiological temperature. Viruses with deletion mutations involving nt 140 to 144 and viruses with large pY1 deletions but normal replication phenotypes in cell culture may have attenuation properties which could be exploited for vaccine development.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andino R., Rieckhof G. E., Baltimore D. A functional ribonucleoprotein complex forms around the 5' end of poliovirus RNA. Cell. 1990 Oct 19;63(2):369–380. doi: 10.1016/0092-8674(90)90170-j. [DOI] [PubMed] [Google Scholar]
- Binn L. N., Lemon S. M., Marchwicki R. H., Redfield R. R., Gates N. L., Bancroft W. H. Primary isolation and serial passage of hepatitis A virus strains in primate cell cultures. J Clin Microbiol. 1984 Jul;20(1):28–33. doi: 10.1128/jcm.20.1.28-33.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown E. A., Day S. P., Jansen R. W., Lemon S. M. The 5' nontranslated region of hepatitis A virus RNA: secondary structure and elements required for translation in vitro. J Virol. 1991 Nov;65(11):5828–5838. doi: 10.1128/jvi.65.11.5828-5838.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown E. A., Zajac A. J., Lemon S. M. In vitro characterization of an internal ribosomal entry site (IRES) present within the 5' nontranslated region of hepatitis A virus RNA: comparison with the IRES of encephalomyocarditis virus. J Virol. 1994 Feb;68(2):1066–1074. doi: 10.1128/jvi.68.2.1066-1074.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang K. H., Brown E. A., Lemon S. M. Cell type-specific proteins which interact with the 5' nontranslated region of hepatitis A virus RNA. J Virol. 1993 Nov;67(11):6716–6725. doi: 10.1128/jvi.67.11.6716-6725.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke B. E., Brown A. L., Currey K. M., Newton S. E., Rowlands D. J., Carroll A. R. Potential secondary and tertiary structure in the genomic RNA of foot and mouth disease virus. Nucleic Acids Res. 1987 Sep 11;15(17):7067–7079. doi: 10.1093/nar/15.17.7067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J. I., Rosenblum B., Ticehurst J. R., Daemer R. J., Feinstone S. M., Purcell R. H. Complete nucleotide sequence of an attenuated hepatitis A virus: comparison with wild-type virus. Proc Natl Acad Sci U S A. 1987 Apr;84(8):2497–2501. doi: 10.1073/pnas.84.8.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J. I., Ticehurst J. R., Feinstone S. M., Rosenblum B., Purcell R. H. Hepatitis A virus cDNA and its RNA transcripts are infectious in cell culture. J Virol. 1987 Oct;61(10):3035–3039. doi: 10.1128/jvi.61.10.3035-3039.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J. I., Ticehurst J. R., Purcell R. H., Buckler-White A., Baroudy B. M. Complete nucleotide sequence of wild-type hepatitis A virus: comparison with different strains of hepatitis A virus and other picornaviruses. J Virol. 1987 Jan;61(1):50–59. doi: 10.1128/jvi.61.1.50-59.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day S. P., Murphy P., Brown E. A., Lemon S. M. Mutations within the 5' nontranslated region of hepatitis A virus RNA which enhance replication in BS-C-1 cells. J Virol. 1992 Nov;66(11):6533–6540. doi: 10.1128/jvi.66.11.6533-6540.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duke G. M., Osorio J. E., Palmenberg A. C. Attenuation of Mengo virus through genetic engineering of the 5' noncoding poly(C) tract. Nature. 1990 Feb 1;343(6257):474–476. doi: 10.1038/343474a0. [DOI] [PubMed] [Google Scholar]
- Escarmís C., Toja M., Medina M., Domingo E. Modifications of the 5' untranslated region of foot-and-mouth disease virus after prolonged persistence in cell culture. Virus Res. 1992 Nov;26(2):113–125. doi: 10.1016/0168-1702(92)90151-x. [DOI] [PubMed] [Google Scholar]
- Gehring K., Leroy J. L., Guéron M. A tetrameric DNA structure with protonated cytosine.cytosine base pairs. Nature. 1993 Jun 10;363(6429):561–565. doi: 10.1038/363561a0. [DOI] [PubMed] [Google Scholar]
- Jacobson A. B., Good L., Simonetti J., Zuker M. Some simple computational methods to improve the folding of large RNAs. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):45–52. doi: 10.1093/nar/12.1part1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang S. K., Davies M. V., Kaufman R. J., Wimmer E. Initiation of protein synthesis by internal entry of ribosomes into the 5' nontranslated region of encephalomyocarditis virus RNA in vivo. J Virol. 1989 Apr;63(4):1651–1660. doi: 10.1128/jvi.63.4.1651-1660.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R. W., Newbold J. E., Lemon S. M. Complete nucleotide sequence of a cell culture-adapted variant of hepatitis A virus: comparison with wild-type virus with restricted capacity for in vitro replication. Virology. 1988 Apr;163(2):299–307. doi: 10.1016/0042-6822(88)90270-x. [DOI] [PubMed] [Google Scholar]
- Kühn R., Luz N., Beck E. Functional analysis of the internal translation initiation site of foot-and-mouth disease virus. J Virol. 1990 Oct;64(10):4625–4631. doi: 10.1128/jvi.64.10.4625-4631.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemon S. M., Binn L. N., Marchwicki R. H. Radioimmunofocus assay for quantitation of hepatitis A virus in cell cultures. J Clin Microbiol. 1983 May;17(5):834–839. doi: 10.1128/jcm.17.5.834-839.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemon S. M., Murphy P. C., Shields P. A., Ping L. H., Feinstone S. M., Cromeans T., Jansen R. W. Antigenic and genetic variation in cytopathic hepatitis A virus variants arising during persistent infection: evidence for genetic recombination. J Virol. 1991 Apr;65(4):2056–2065. doi: 10.1128/jvi.65.4.2056-2065.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowman H. B., Draper D. E. On the recognition of helical RNA by cobra venom V1 nuclease. J Biol Chem. 1986 Apr 25;261(12):5396–5403. [PubMed] [Google Scholar]
- Paul A. V., Tada H., von der Helm K., Wissel T., Kiehn R., Wimmer E., Deinhardt F. The entire nucleotide sequence of the genome of human hepatitis A virus (isolate MBB). Virus Res. 1987 Aug;8(2):153–171. doi: 10.1016/0168-1702(87)90026-8. [DOI] [PubMed] [Google Scholar]
- Pilipenko E. V., Gmyl A. P., Maslova S. V., Svitkin Y. V., Sinyakov A. N., Agol V. I. Prokaryotic-like cis elements in the cap-independent internal initiation of translation on picornavirus RNA. Cell. 1992 Jan 10;68(1):119–131. doi: 10.1016/0092-8674(92)90211-t. [DOI] [PubMed] [Google Scholar]
- Rieder E., Bunch T., Brown F., Mason P. W. Genetically engineered foot-and-mouth disease viruses with poly(C) tracts of two nucleotides are virulent in mice. J Virol. 1993 Sep;67(9):5139–5145. doi: 10.1128/jvi.67.9.5139-5145.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whetter L. E., Day S. P., Elroy-Stein O., Brown E. A., Lemon S. M. Low efficiency of the 5' nontranslated region of hepatitis A virus RNA in directing cap-independent translation in permissive monkey kidney cells. J Virol. 1994 Aug;68(8):5253–5263. doi: 10.1128/jvi.68.8.5253-5263.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]