Abstract
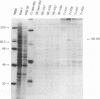
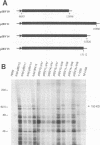
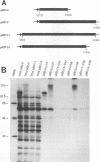
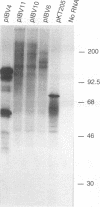
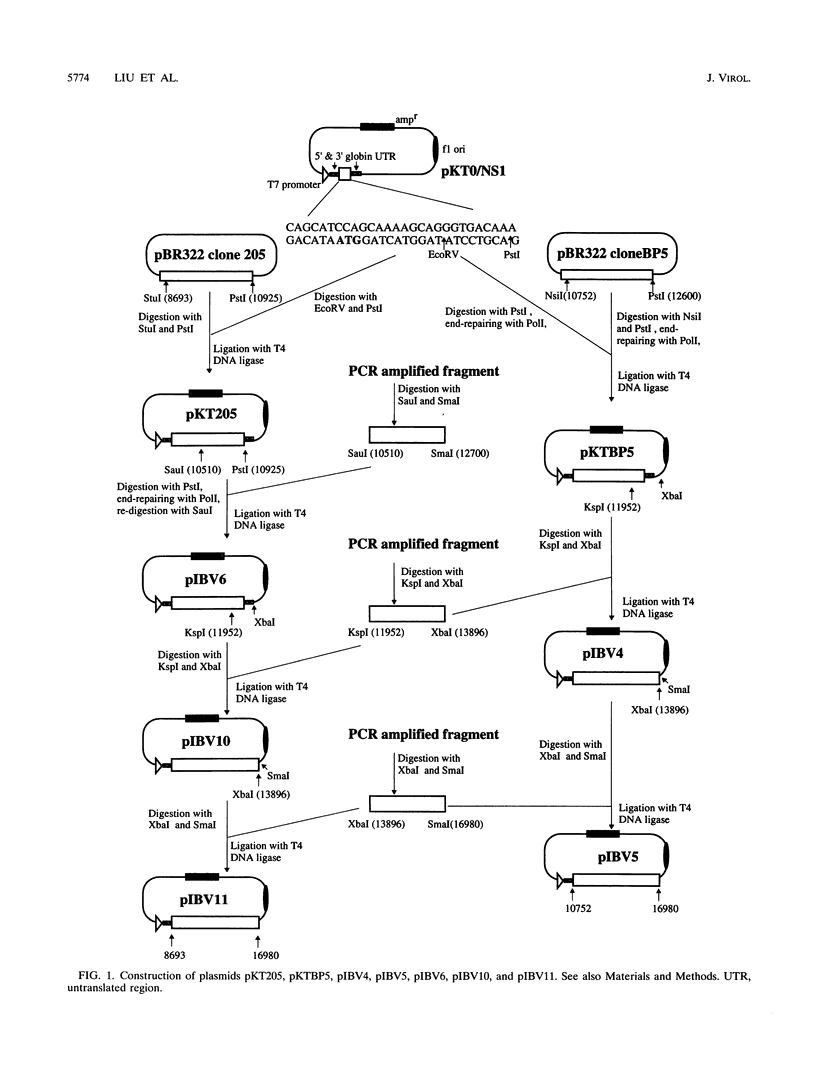
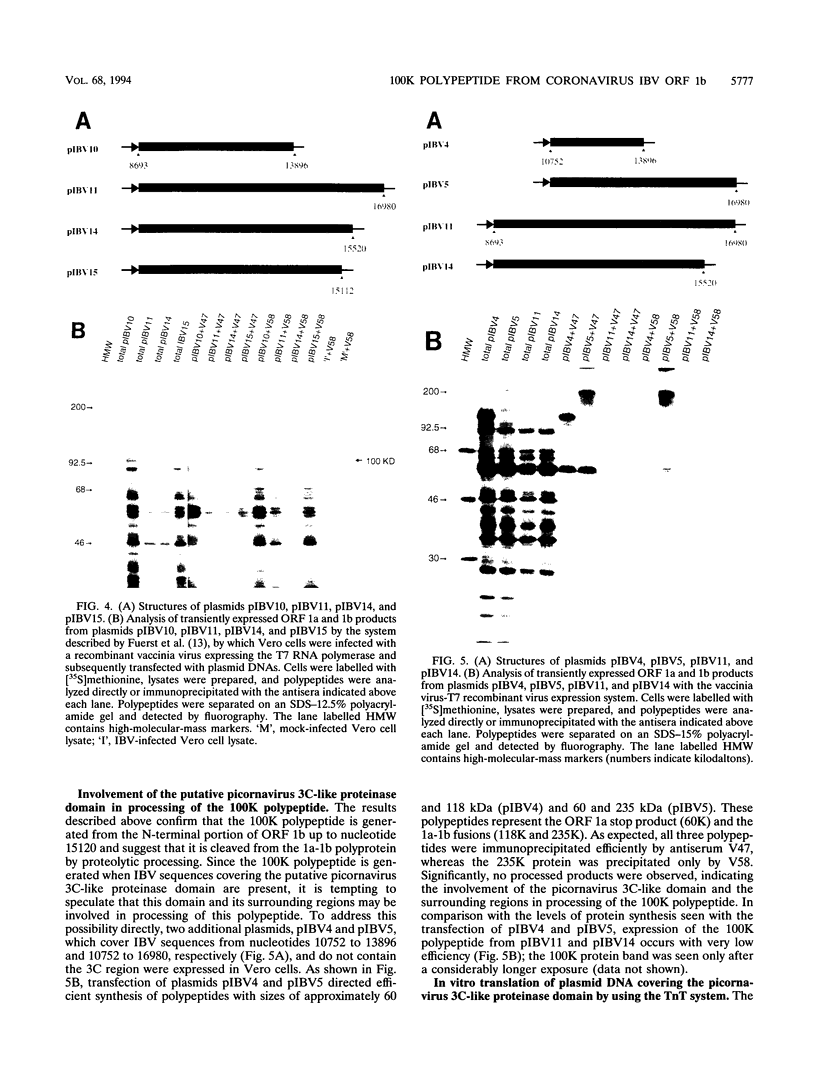
The genome-length mRNA (mRNA 1) of the coronavirus infectious bronchitis virus (IBV) contains two large open reading frames (ORFs), 1a and 1b, with the potential to encode polypeptides of 441 and 300 kDa, respectively. The downstream ORF, ORF 1b, is expressed by a ribosomal frameshifting mechanism. In an effort to detect viral polypeptides encoded by ORF 1b in virus-infected cells, immunoprecipitations were carried out with a panel of region-specific antisera. A polypeptide of approximately 100 kDa was precipitated from IBV-infected, but not mock-infected, Vero cells by one of these antisera (V58). Antiserum V58 was raised against a bacterially expressed fusion protein containing polypeptide sequences encoded by ORF 1b nucleotides 14492 to 15520; it recognizes specifically the corresponding in vitro-synthesized target protein. A polypeptide comigrating with the 100,000-molecular-weight protein (100K protein) identified in infected cells was also detected when the IBV sequence from nucleotides 8693 to 16980 was expressed in Vero cells by using a vaccinia virus-T7 expression system. Deletion analysis revealed that the sequence encoding the C terminus of the 100K polypeptide lies close to nucleotide 15120; it may therefore be generated by proteolysis at a potential QS cleavage site encoded by nucleotides 15129 to 15135. In contrast, expression of IBV sequences from nucleotides 10752 to 16980 generated two polypeptides of approximately 62 and 235 kDa, which represent the ORF 1a stop product and the 1a-1b fused product generated by a frameshifting mechanism, respectively, but no processed products were observed. Since the putative picornavirus 3C-like proteinase domain is located in ORF 1a between nucleotides 8937 and 9357, this observation suggests that deletion of the picornavirus 3C-like proteinase domain and surrounding regions abolishes processing of the 1b polyprotein. In addition, the in vitro translation and in vivo transfection studies also indicate that the ORF 1a region between nucleotides 8763 and 10720 contains elements that down-regulate the expression of ORF 1b.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baker S. C., Shieh C. K., Soe L. H., Chang M. F., Vannier D. M., Lai M. M. Identification of a domain required for autoproteolytic cleavage of murine coronavirus gene A polyprotein. J Virol. 1989 Sep;63(9):3693–3699. doi: 10.1128/jvi.63.9.3693-3699.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker S. C., Yokomori K., Dong S., Carlisle R., Gorbalenya A. E., Koonin E. V., Lai M. M. Identification of the catalytic sites of a papain-like cysteine proteinase of murine coronavirus. J Virol. 1993 Oct;67(10):6056–6063. doi: 10.1128/jvi.67.10.6056-6063.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boursnell M. E., Brown T. D., Foulds I. J., Green P. F., Tomley F. M., Binns M. M. Completion of the sequence of the genome of the coronavirus avian infectious bronchitis virus. J Gen Virol. 1987 Jan;68(Pt 1):57–77. doi: 10.1099/0022-1317-68-1-57. [DOI] [PubMed] [Google Scholar]
- Bredenbeek P. J., Pachuk C. J., Noten A. F., Charité J., Luytjes W., Weiss S. R., Spaan W. J. The primary structure and expression of the second open reading frame of the polymerase gene of the coronavirus MHV-A59; a highly conserved polymerase is expressed by an efficient ribosomal frameshifting mechanism. Nucleic Acids Res. 1990 Apr 11;18(7):1825–1832. doi: 10.1093/nar/18.7.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I., Boursnell M. E., Binns M. M., Bilimoria B., Blok V. C., Brown T. D., Inglis S. C. An efficient ribosomal frame-shifting signal in the polymerase-encoding region of the coronavirus IBV. EMBO J. 1987 Dec 1;6(12):3779–3785. doi: 10.1002/j.1460-2075.1987.tb02713.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I., Boursnell M. E., Binns M. M., Bilimoria B., Rolley N. J., Brown T. D., Inglis S. C. Products of the polymerase-encoding region of the coronavirus IBV. Adv Exp Med Biol. 1990;276:275–281. doi: 10.1007/978-1-4684-5823-7_38. [DOI] [PubMed] [Google Scholar]
- Brierley I., Digard P., Inglis S. C. Characterization of an efficient coronavirus ribosomal frameshifting signal: requirement for an RNA pseudoknot. Cell. 1989 May 19;57(4):537–547. doi: 10.1016/0092-8674(89)90124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh D., Brian D. A., Enjuanes L., Holmes K. V., Lai M. M., Laude H., Siddell S. G., Spaan W., Taguchi F., Talbot P. J. Recommendations of the Coronavirus Study Group for the nomenclature of the structural proteins, mRNAs, and genes of coronaviruses. Virology. 1990 May;176(1):306–307. doi: 10.1016/0042-6822(90)90259-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras R., Cheroutre H., Degrave W., Fiers W. Simple, efficient in vitro synthesis of capped RNA useful for direct expression of cloned eukaryotic genes. Nucleic Acids Res. 1982 Oct 25;10(20):6353–6362. doi: 10.1093/nar/10.20.6353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denison M. R., Perlman S. Translation and processing of mouse hepatitis virus virion RNA in a cell-free system. J Virol. 1986 Oct;60(1):12–18. doi: 10.1128/jvi.60.1.12-18.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denison M. R., Zoltick P. W., Hughes S. A., Giangreco B., Olson A. L., Perlman S., Leibowitz J. L., Weiss S. R. Intracellular processing of the N-terminal ORF 1a proteins of the coronavirus MHV-A59 requires multiple proteolytic events. Virology. 1992 Jul;189(1):274–284. doi: 10.1016/0042-6822(92)90703-R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denison M. R., Zoltick P. W., Leibowitz J. L., Pachuk C. J., Weiss S. R. Identification of polypeptides encoded in open reading frame 1b of the putative polymerase gene of the murine coronavirus mouse hepatitis virus A59. J Virol. 1991 Jun;65(6):3076–3082. doi: 10.1128/jvi.65.6.3076-3082.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuerst T. R., Niles E. G., Studier F. W., Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci U S A. 1986 Nov;83(21):8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gay D. A., Yen T. J., Lau J. T., Cleveland D. W. Sequences that confer beta-tubulin autoregulation through modulated mRNA stability reside within exon 1 of a beta-tubulin mRNA. Cell. 1987 Aug 28;50(5):671–679. doi: 10.1016/0092-8674(87)90325-4. [DOI] [PubMed] [Google Scholar]
- Gorbalenya A. E., Koonin E. V., Donchenko A. P., Blinov V. M. Coronavirus genome: prediction of putative functional domains in the non-structural polyprotein by comparative amino acid sequence analysis. Nucleic Acids Res. 1989 Jun 26;17(12):4847–4861. doi: 10.1093/nar/17.12.4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold J., Raabe T., Schelle-Prinz B., Siddell S. G. Nucleotide sequence of the human coronavirus 229E RNA polymerase locus. Virology. 1993 Aug;195(2):680–691. doi: 10.1006/viro.1993.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lee H. J., Shieh C. K., Gorbalenya A. E., Koonin E. V., La Monica N., Tuler J., Bagdzhadzhyan A., Lai M. M. The complete sequence (22 kilobases) of murine coronavirus gene 1 encoding the putative proteases and RNA polymerase. Virology. 1991 Feb;180(2):567–582. doi: 10.1016/0042-6822(91)90071-I. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D. X., Cavanagh D., Green P., Inglis S. C. A polycistronic mRNA specified by the coronavirus infectious bronchitis virus. Virology. 1991 Oct;184(2):531–544. doi: 10.1016/0042-6822(91)90423-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D. X., Gompels U. A., Nicholas J., Lelliott C. Identification and expression of the human herpesvirus 6 glycoprotein H and interaction with an accessory 40K glycoprotein. J Gen Virol. 1993 Sep;74(Pt 9):1847–1857. doi: 10.1099/0022-1317-74-9-1847. [DOI] [PubMed] [Google Scholar]
- Liu D. X., Inglis S. C. Identification of two new polypeptides encoded by mRNA5 of the coronavirus infectious bronchitis virus. Virology. 1992 Jan;186(1):342–347. doi: 10.1016/0042-6822(92)90094-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D. X., Inglis S. C. Internal entry of ribosomes on a tricistronic mRNA encoded by infectious bronchitis virus. J Virol. 1992 Oct;66(10):6143–6154. doi: 10.1128/jvi.66.10.6143-6154.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberst M. D., Gollan T. J., Gupta M., Peura S. R., Zydlewski J. D., Sudarsanan P., Lawson T. G. The encephalomyocarditis virus 3C protease is rapidly degraded by an ATP-dependent proteolytic system in reticulocyte lysate. Virology. 1993 Mar;193(1):28–40. doi: 10.1006/viro.1993.1100. [DOI] [PubMed] [Google Scholar]
- Pachter J. S., Yen T. J., Cleveland D. W. Autoregulation of tubulin expression is achieved through specific degradation of polysomal tubulin mRNAs. Cell. 1987 Oct 23;51(2):283–292. doi: 10.1016/0092-8674(87)90155-3. [DOI] [PubMed] [Google Scholar]
- Parker R., Jacobson A. Translation and a 42-nucleotide segment within the coding region of the mRNA encoded by the MAT alpha 1 gene are involved in promoting rapid mRNA decay in yeast. Proc Natl Acad Sci U S A. 1990 Apr;87(7):2780–2784. doi: 10.1073/pnas.87.7.2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyu A. B., Greenberg M. E., Belasco J. G. The c-fos transcript is targeted for rapid decay by two distinct mRNA degradation pathways. Genes Dev. 1989 Jan;3(1):60–72. doi: 10.1101/gad.3.1.60. [DOI] [PubMed] [Google Scholar]
- Smith A. R., Boursnell M. E., Binns M. M., Brown T. D., Inglis S. C. Identification of a new membrane-associated polypeptide specified by the coronavirus infectious bronchitis virus. J Gen Virol. 1990 Jan;71(Pt 1):3–11. doi: 10.1099/0022-1317-71-1-3. [DOI] [PubMed] [Google Scholar]
- Stern D. F., Kennedy S. I. Coronavirus multiplication strategy. I. Identification and characterization of virus-specified RNA. J Virol. 1980 Jun;34(3):665–674. doi: 10.1128/jvi.34.3.665-674.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern D. F., Kennedy S. I. Coronavirus multiplication strategy. II. Mapping the avian infectious bronchitis virus intracellular RNA species to the genome. J Virol. 1980 Nov;36(2):440–449. doi: 10.1128/jvi.36.2.440-449.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern D. F., Sefton B. M. Coronavirus multiplication: locations of genes for virion proteins on the avian infectious bronchitis virus genome. J Virol. 1984 Apr;50(1):22–29. doi: 10.1128/jvi.50.1.22-29.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]