Abstract
The retinoids are reported to reduce incidence of second primary aerodigestive cancers. Mechanisms for this chemoprevention are previously linked to all-trans retinoic acid (RA) signaling growth inhibition at G1 in carcinogen-exposed immortalized human bronchial epithelial cells. This study investigated how RA suppresses human bronchial epithelial cell growth at the G1-S cell cycle transition. RA signaled growth suppression of human bronchial epithelial cells and a decline in cyclin D1 protein but not mRNA expression. Exogenous cyclin D1 protein also declined after RA treatment of transfected, immortalized human bronchial epithelial cells, suggesting that posttranslational mechanisms were active in this regulation of cyclin D1 expression. Findings were extended by showing treatment with ubiquitin-dependent proteasome inhibitors: calpain inhibitor I and lactacystin each prevented this decreased cyclin D1 protein expression, despite RA treatment. Treatment with the cysteine proteinase inhibitor, E-64, did not prevent this cyclin D1 decline. High molecular weight cyclin D1 protein species appeared after proteasome inhibitor treatments, suggesting that ubiquitinated species were present. To learn whether RA directly promoted degradation of cyclin D1 protein, studies using human bronchial epithelial cell protein extracts and in vitro-translated cyclin D1 were performed. In vitro-translated cyclin D1 degraded more rapidly when incubated with extracts from RA treated vs. untreated cells. Notably, this RA-signaled cyclin D1 proteolysis depended on the C-terminal PEST sequence, a region rich in proline (P), glutamate (E), serine (S), and threonine (T). Taken together, these data highlight RA-induced cyclin D1 proteolysis as a mechanism signaling growth inhibition at G1 active in the prevention of human bronchial epithelial cell transformation.
The retinoids are natural and synthetic analogs of vitamin A. Retinoids are reported to treat oral leukoplakia (1) and to reduce second primary hepatocellular or aerodigestive tract cancers (2–4). The mechanisms responsible for this reduction of second primary cancers are poorly understood. We previously reported that all-trans retinoic acid (RA) inhibits carcinogen-induced transformation of human bronchial epithelial cells and that this is linked to a delayed G1-S cell cycle transition (5). It was hypothesized that RA protects cells from carcinogen-induced transformation by permitting repair of mutagenized genomic DNA before subsequent rounds of cell division. The current study examined how RA regulates expression of the G1 cyclin, cyclin D1.
Cell cycle transition occurs through activation and inactivation of cyclin-dependent kinases (Cdks). Cdks become activated by complexing with specific cyclins expressed during the cell cycle (6, 7). Cyclin–Cdk complexes are inhibited by the binding of specific cyclin inhibitors (8). In eukaryotic cells, cyclin D expression increases in mid-G1, complexing to Cdk4 and Cdk6 and producing peak activation near the G1-S cell cycle transition (6, 7, 9, 10). Cyclin E expression increases in late G1, complexing to and activating Cdk2 (10–13). Expression of cyclin A accumulates during S and G2 phases, and expression of cyclin B is typically maximal during the G2-M cell cycle transition (6, 7).
Cyclin proteolysis is essential for cell cycle progression, as recently reviewed (14, 15). Cyclins E, A, and B are regulated by a ubiquitin-dependent degradation pathway (14–16). Ubiquitin is a 76-amino acid polypeptide highly conserved in eukaryotic cells (17). It is activated in an ATP-dependent manner by a thiol ester link to a ubiquitin-activating enzyme, E1 (18). Activated ubiquitin is then bound to the conjugating enzyme, E2 (18, 19). Ubiquitin is transferred to specific proteins by E2, often requiring an E3 ligase (20, 21). Subsequent attachment of ubiquitin monomers to the substrates results in multi-ubiquitinated chains degraded by the 26S proteasome (15, 22).
This study reports that RA directly signals a decline in cyclin D1 protein expression in human bronchial epithelial cells through induced proteolysis. The ubiquitin-dependent proteasome degradation pathway is implicated in this retinoid effect. RA-signaled cyclin D1 proteolysis is proposed as a mechanism linked to growth suppression during prevention of human bronchial epithelial cell transformation.
MATERIALS AND METHODS
Cell Lines, Culture Conditions, and Expression Vectors.
The proteasome inhibitors calpain inhibitor I (Calbiochem–Nova Biochem) and lactacystin (23) were used. BEAS-2B cells were derived from normal human bronchial epithelial cells immortalized with an adenovirus 12-simian virus 40 hybrid virus (24). BEAS-2B cells were cultured in serum free medium, as described (25). To construct the Eboplpp–cyclin D1 expression vector, the BamHI–EcoRI 1.1-kb full length cDNA fragment of murine cyclin D1 was isolated from the pGEX-3X-cyclin D1 plasmid and cloned into the Eboplpp vector (26) at the HindIII restriction endonuclease site. The murine cyclin D1 cDNA was cloned into the EcoRV site of the Bluescript II plasmid (Stratagene). Transfections were performed using lipofectamine (GIBCO/BRL) and selection in hygromycin (40 μg/ml), as described (5).
Immunoblot Analysis.
BEAS-2B cells were lysed in a modified radioimmunoprecipitation assay buffer (5), and protein concentrations were measured using the Bradford assay. Total cellular protein was size-fractionated by SDS/PAGE, then transferred to a nitrocellulose filter (Schleicher & Schuell) before incubation with the desired primary antibody and protein detection using the chemiluminescence assay (Amersham). The anti-human cyclin D1 (M20), anti-murine cyclin D1 (72-13G), and anti-human cyclin E (HE 12) antibodies were purchased from Santa Cruz Biotechnology.
Proliferation Assay.
Cell proliferation was measured by a H3-thymidine incorporation assay. In brief, 105 BEAS-2B cells were plated in duplicate wells of a 6-well tissue culture plate and treated with RA (2 μM or 4 μM) or dimethyl sulfoxide (DMSO, 1:10,000 dilution) for 3, 6, and 24 h. At these time points, the cells were treated for 1 h with 4 μCi/ml of H3-thymidine, washed with PBS, and lysed (10 mM sodium hydroxide/1% SDS), and H3-thymidine incorporation was measured using a scintillation counter.
Northern Analysis.
Ten micrograms of total cellular RNA were size-fractionated on a 1% agarose–formaldehyde gel before transfer to a Hybond-N membrane (Amersham). Membranes were incubated overnight at 66°C with 200 μg/ml digoxin-labeled anti-sense cyclin D1 riboprobe. Membranes were stringently washed, and mRNA expression was detected per manufacturer’s recommendations. (Boehringer Mannheim).
Immunoprecipitation.
BEAS-2B cells were lysed with modified radioimmunoprecipitation assay buffer (5), and a monoclonal anti-cyclin D1 antibody (HD11, Santa Cruz) was used for immunoprecipitation using established techniques (27). Replicate immunoblots were incubated with a second polyclonal anti-cyclin D1 antibody (M20, Santa Cruz) or an anti-ubiquitin antibody.
In Vitro Translation of Cyclin D1.
Full length cyclin D1 mRNA was transcribed from the described Bluescript plasmid containing cyclin D1 using the T7 promoter (27). To remove the PEST sequence, this plasmid was linearized 76 bp proximal to the 3′ end of the cyclin D1 cDNA. Cyclin D1 protein was in vitro translated using 1 μg of transcribed mRNA added to 35 μl of rabbit reticulocyte lysates (28) containing S35-methionine (Promega) at 25°C for 1.5 h.
In Vitro Degradation of Cyclin D1.
BEAS-2B cellular extracts were isolated by scraping cells in ice-cold PBS containing 1 mM MgCl2 before centrifugation. Cell pellets were lysed in two original volumes of hypotonic buffer (10 mM Tris, pH 8.3) for 10 minutes on ice and briefly sonicated (29), and the supernatant was collected after centrifugation. The reaction buffer contained 10 mM Tris, 5 mM CaCl2, 5 mM MgCl2 (pH 7.5), an ATP regeneration system (3 units/ml creatine phosphokinase, 10 mM phosphocreatine, and 2 mM ATP) and 1 mM of the protein synthesis inhibitor emetine. Reactions were performed in 150-μl aliquots containing 33% vol/vol of cellular extract at a final protein concentration of 2.3 μg/μl. Rabbit reticulocyte lysates containing radiolabeled cyclin D1 were added at 2% vol/vol. The reaction mixtures were incubated at 37°C, and aliquots were removed at desired time points before terminating reactions in SDS running buffer. Samples were subjected to SDS/PAGE, and the S35-methionine incorporation was quantified using a PhosphorImager (Molecular Dynamics).
RESULTS
RA Represses Cyclin D1 Protein and Human Bronchial Epithelial Cell Growth.
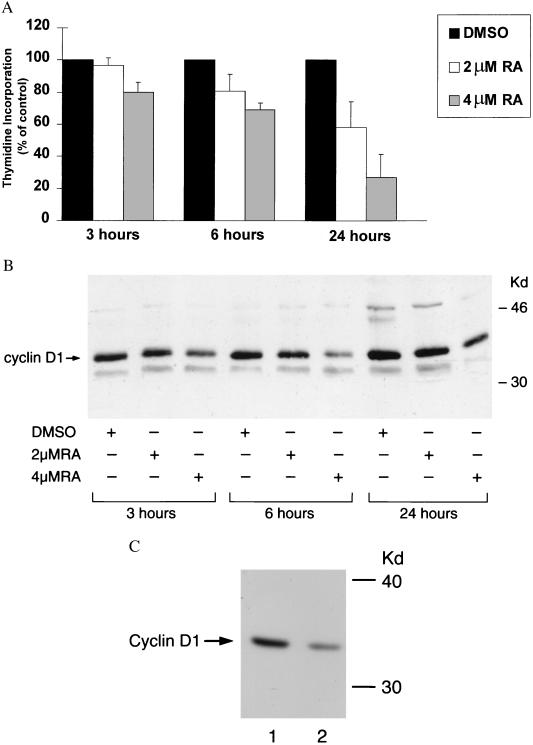
RA-mediated changes in G1-S cyclin expression and cellular proliferation were studied using the immortalized human bronchial epithelial cell line BEAS-2B (Fig. 1 A and B). Immunoblot findings revealed that a dose-dependent decline in cyclin D1 expression follows RA treatment of BEAS-2B cells. Cyclin D1 protein levels were maximally suppressed 6–12 h after RA treatment. Cyclin E expression also decreased after RA treatment, typically 3–6 h after the observed decline in cyclin D1 (data not shown). After RA treatment, H3-thymidine incorporation decreased concomitantly with the observed decline in cyclin D1 expression. Cultured normal human bronchial epithelial cells were also examined for RA-mediated changes in cyclin D1 protein expression. These RA-treated normal human bronchial epithelial cells also decreased cyclin D1 protein expression relative to controls (Fig. 1C). Normal human bronchial epithelial cells responded to RA treatment (4 μM) compared with vehicle controls with a 45% decrease in thymidine incorporation signaled by 6 h (data not shown).
Figure 1.
A decline in growth and cyclin D1 protein expression follows RA treatment of human bronchial epithelial cells. Exponentially growing BEAS-2B cells were treated with DMSO vehicle, 2 μM RA, or 4 μM RA for 3, 6, and 24 h and (A) H3-thymidine incorporation was measured. Using protein isolated from these cells, (B) cyclin D1 immunoblots were performed as described in Materials and Methods. Repressed BEAS-2B growth and cyclin D1 expression occurred in the first 3–6 h after RA treatment. (C) Immunoblot expression for cyclin D1 in DMSO vehicle-treated (lane 1) vs. RA-treated (4 μM RA, lane 2) normal human bronchial epithelial cells reveals that RA treatment markedly represses cyclin D1 expression in these cells.
RA Decreases Exogenous Cyclin D1 Expression.
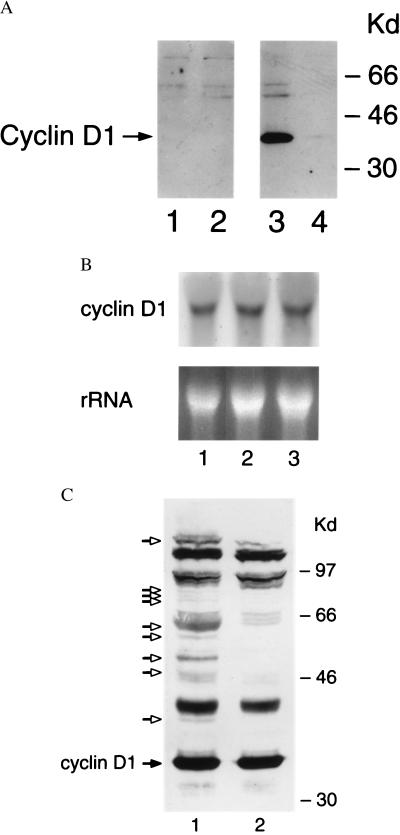
Endogenous cyclin D1 expression is markedly repressed by RA treatment. Whether exogenously expressed cyclin D1 also responds to RA treatment was studied. A simian virus 40-driven murine cyclin D1 cDNA engineered in the Eboplpp episomal expression vector was overexpressed in BEAS-2B cells. These BEAS-2B transfectants remained sensitive to the growth suppressive effects of RA (data not shown). This is likely caused by RA treatment repressing exogenous cyclin D1 expression, as shown by Western analysis using a murine-specific anti-cyclin D1 antibody (Fig. 2A). Cyclin D1 mRNA expression was examined at 3, 6, and 24 h after treatment with RA or vehicle control (Fig. 2B and data not shown). RA treatment of BEAS-2B cells, compared with vehicle controls, did not repress cyclin D1 mRNA expression. These data implicated posttranscriptional mechanisms in the RA-signaled repression of cyclin D1.
Figure 2.
RA treatment signals a decline in cyclin D1 protein but not mRNA expression in BEAS-2B cells. (A) BEAS-2B cells transfected with the insertless Eboplpp vector (lanes 1 and 2) or with murine cyclin D1 (lanes 3 and 4) were treated with DMSO vehicle (lanes 1 and 3) or with 4 μM RA (lanes 2 and 4) for 6 h. Immunoblots were probed with a murine-specific anti-cyclin D1 antibody. RA treatment represses exogenous cyclin D1 expression. (B) Northern analysis for cyclin D1 expression was performed as described in the Materials and Methods. Cyclin D1 mRNA expression in BEAS-2B cells was analyzed at 6 h after treatment with DMSO vehicle (lane 1), 2 μM RA (lane 2), or 4 μM RA (lane 3). Cyclin D1 mRNA expression was unaffected by RA treatment. Similar findings were observed at 3 and 24 h after RA treatment (data not shown). Ethidium-stained ribosomal bands rRNA are provided to confirm that similar amounts of total RNA are added per lane. (C) The posttranslational modification of cyclin D1 in BEAS-2B cells. From BEAS-2B cells treated with the calpain inhibitor I (lane 1) or the DMSO vehicle (lane 2) for 24 h, 400 μg of total cellular protein was size-fractionated by SDS/PAGE as described in Materials and Methods. This cyclin D1 immunoblot revealed cyclin D1 (closed arrow) and high molecular weight cyclin D1 reactive species (open arrows) in the calpain-treated cells.
Posttranslational Modification of Cyclin D1.
Eukaryotic cyclins E, A, and B are tightly regulated via induction of protein proteolysis by ubiquitin-dependent pathways (14–16). Whether cyclin D1 is also regulated by the ubiquitin degradation pathway in BEAS-2B cells was studied. BEAS-2B cells treated with the 26S proteasome inhibitor calpain inhibitor I for 24 h exhibited accumulation of high molecular weight cyclin D1 immunoreactive species as shown in Fig. 2C. Some of these high molecular weight species were immunoreactive both with a second anti-cyclin D1 antibody and an anti-ubiquitin antibody (data not shown). These high molecular weight species were undetected when an antibody that does not identify cyclin D1 was used in this assay, and these species were undetected when a blocking cyclin D1 peptide was incubated with the anti-cyclin D1 antisera (data not shown).
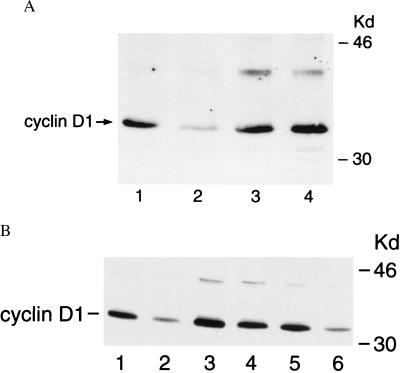
Proteasome Involvement in RA-Mediated Decline of Cyclin D1.
We next examined whether blocking the 26S proteasome antagonizes RA-mediated decline in cyclin D1 expression. BEAS-2B cells, treated with calpain inhibitor I (100 μM) for 1.5 h before RA treatment (4 μM) for 6 h, prevented the expected decline in cyclin D1 protein expression (Fig. 3A). This finding was confirmed and extended using another proteasome inhibitor, lactacystin, which also prevented the expected decline in cyclin D1 expression by RA treatment (Fig. 3B). Notably, the cysteine proteinase inhibitor, E-64, did not inhibit this decline of cyclin D1 after RA treatment as shown in Fig. 3B. These data are consistent with the view that RA treatment represses cyclin D1 expression by promoting proteolysis via ubiquitin-dependent pathways.
Figure 3.
Inhibition of the 26S proteasome pathway-antagonized, RA-mediated decline in cyclin D1. BEAS-2B cells were treated with the indicated proteasome inhibitors or the cysteine proteinase inhibitor (E-64) for 1.5 h before incubation with DMSO vehicle or 4 μM RA for 6 h. Cyclin D1 immunoblots were performed for (A) BEAS-2B cells treated with vehicle (lane 1) or 4 μM RA (lane 2) or treated with calpain inhibitor I (100 μM) and vehicle (lane 3) or calpain inhibitor I (100 μM) and 4 μM RA (lane 4). (B) BEAS-2B cells were treated with vehicle (lane 1) or 4 μM RA (lane 2) or treated with lactacystin (100 μM) and vehicle (lane 3) or lactacystin (100 μM) and 4 μM RA (lane 4) or treated with E-64 (100 μM) and vehicle (lane 5) or E-64 (100 μM) and 4 μM RA (lane 6). The proteasome inhibitors but not the cysteine proteinase inhibitor blocked RA-mediated decline in cyclin D1 expression.
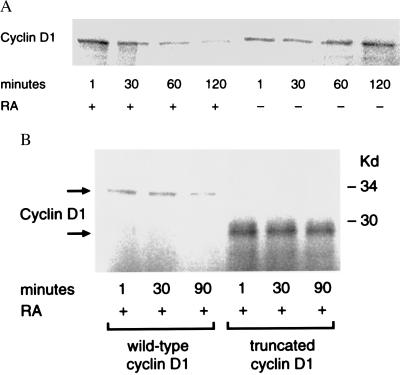
RA Promotes Degradation of Cyclin D1 Protein.
To confirm that RA regulated cyclin D1 expression by activating cyclin D1 proteolysis, the degradation of cyclin D1 was studied in a cell-free system. Cellular protein extracts were obtained from exponentially growing BEAS-2B cells treated with RA (4 μM) or DMSO vehicle control. Radiolabeled cyclin D1 degraded more rapidly in BEAS-2B cellular extracts obtained from RA-treated compared with controls cells (Fig. 4A). Removal of the C-terminal PEST sequence of radiolabeled cyclin D1 prevented this degradation by cell extracts derived from RA-treated cells (Fig. 4B). S35 methionine pulse–chase experiments revealed degradation of cyclin D1 protein in vivo was augmented in BEAS-2B cells treated with RA (4 μM) compared with DMSO controls (data not shown).
Figure 4.
RA promotes degradation of cyclin D1 in vitro. In vitro-translated, S35-methionine-labeled cyclin D1 was incubated with cell extracts from BEAS-2B cells treated with DMSO vehicle or 4 μM RA for 4.5 h. Protein aliquots were assayed by SDS/PAGE and expression detected with a PhosphorImager, as described in Materials and Methods. (A) In vitro-translated cyclin D1 degraded more rapidly when exposed to RA-treated (+) compared with vehicle control (−) BEAS-2B cellular extracts. (B) Degradation of radiolabeled wild-type cyclin D1 by RA (upper arrow) is antagonized (lower arrow) by removal of the C-terminal PEST sequence (truncated cyclin D1).
DISCUSSION
Retinoids are reported to prevent second primary aerodigestive tract and hepatocellular carcinomas and to treat the premalignant lesion oral leukoplakia (2–4). Mechanisms responsible for these beneficial clinical effects are not yet known. This study reports that RA decreases cyclin D1 protein expression paralleling a decline in proliferation of human bronchial epithelial cells. RA also signals a decline in cyclin E expression (5) after repressed cyclin D1 expression. Because cyclins E and D1 are viewed as rate-limiting in the G1-S cell cycle transition, the data presented here are consistent with RA-suppressing cell growth directly through repressed cyclin D1 and/or cyclin E expression.
This study reveals that RA signals cyclin D1 proteolysis. Cyclin D1 mRNA is not regulated by RA treatment of immortalized human bronchial epithelial cells. Expression of exogenous cyclin D1 protein in BEAS-2B cells also was repressed by RA treatment, as shown in Fig. 2. Findings were extended by demonstrating that RA enhances degradation of cyclin D1 in vitro and in vivo (Figs. 1 and 4). These data are consistent with a posttranscriptional mechanism active in this degradation and point to a role for the ubiquitin pathway in this cyclin D1 proteolysis signaled by RA treatment. Notably, the proteasome inhibitors calpain inhibitor I and lactacystin each prevented cyclin D1 repression by RA treatment, as depicted in Fig. 3. In a cell-free system, cyclin D1 degradation by RA depended on the C-terminal PEST sequence as shown in Fig. 4. This C-terminal region of cyclin D1 contains the PEST sequence. Recent reports demonstrate that the phosphorylation of a single C-terminal threonine in the PEST sequence of cyclins D1 and E regulates ubiquitination (30, 31). Thus, the C-terminal PEST region in these G1 cyclins appears involved in ubiquitination and degradation. Cyclins A and B do not contain the PEST sequence (32) but require the amino-terminal destruction box for ubiquitin degradation. The link between the ubiquitin–proteasome pathway and RA-regulated cyclin expression reported here is reminiscent of the decline of PML/retinoic acid receptor-α by RA treatment of acute promyleocytic leukemia cells through a ubiquitin-dependent pathway (33).
Although cyclin E is repressed by RA treatment, it is not yet known whether RA enhances cyclin E proteolysis. Preliminary findings suggest that cyclin E also is repressed by RA through mechanisms similar to those active in cyclin D1 degradation. RA does not appear to affect cyclin E mRNA expression in BEAS-2B cells (data not shown). Overexpression of cyclin E in immortalized human bronchial epithelial cells does not antagonize retinoid growth inhibition (5). It will be interesting to determine whether RA signals a general induction of the ubiquitin pathway or specifically targets G1-S cyclins. Future work will clarify how RA promotes cyclin D1 degradation and whether RA affects either Thr-286 phosphorylation, ubiquitin conjugation, or enhanced proteasome degradation of cyclin D1.
The findings presented here are notable because this report directly links retinoid growth suppression at G1 to a specific proteolysis mechanism. These data reveal that RA signals cyclin D1 proteolysis and strongly implicate involvement of the ubiquitin pathway in this regulation. Regulating cyclin expression has clinical relevancy because aberrant expression of cyclins D1 and E is frequent in human tumors (34–37). In this regard, retinoid targeting of cyclin D1 degradation represents a chemoprevention mechanism having therapeutic implications for aerodigestive tract tumors.
Acknowledgments
We thank Dr. Curtis Harris, Laboratory of Human Carcinogenesis, National Cancer Institute, for the gift of the BEAS-2B cell line, Dr. Charles Sherr, St. Jude Children’s Research Hospital, for pGEX-3X-cyclin D1, Dr. Satoshi Omura, The Kitasato Institute, Japan, for the gift of lactacystin, and Dr. Arthur L. Haas, Medical College of Wisconsin, for the gift of anti-ubiquitin antibody. We thank Dr. Andrew Koff, Molecular Biology Program, Sloan–Kettering Institute and Dr. Michele Pagano, New York University, for helpful consultations. This work was supported by a research grant from Pfizer, by National Institutes of Health Grant RO1 CA54494–04 (E.D.), and by the ASCO Young Investigator Award (J.B.). J.L. was supported by National Institutes of Health Grants T32-CA09512 and K12-CA01712, and J.B. was supported by National Institutes of Health Grant T32-CA09685 from the National Cancer Institute. H.K. was supported by the DeWitt Wallace Research Fund of Memorial Sloan–Kettering Cancer Center.
ABBREVIATIONS
- RA
all-trans retinoic acid
- Cdk
cyclin-dependent kinase
- DMSO
dimethyl sulfoxide
- PEST
a region rich in proline (P), glutamate (E), serine (S), and threonine (T)
References
- 1.Hong W K, Endicott J, Itri L M, Doos W, Batsakis J G, Bell R, Fofonoff S, Byers R, Atkinson E N, Vaughan C, Toth B B, Kramer A, Dimery I W, Skipper P, Strong S. N Engl J Med. 1986;315:1501–1505. doi: 10.1056/NEJM198612113152401. [DOI] [PubMed] [Google Scholar]
- 2.Hong W K, Lippman S M, Itri L M, Karp D D, Lee J S, Byers R M, Schantz S P, Kramer A M, Lotan R, Peters L J, Dimery I W, Brown B W, Goepfert H. N Engl J Med. 1990;323:795–801. doi: 10.1056/NEJM199009203231205. [DOI] [PubMed] [Google Scholar]
- 3.Pastorino U, Infante M, Maioli M, Chiesa G, Buyse M, Firket P, Rosmentz N, Clerici M, Soresi E, Valente M, Belloni P A, Ravasi G. J Clin Oncol. 1993;11:1216–1222. doi: 10.1200/JCO.1993.11.7.1216. [DOI] [PubMed] [Google Scholar]
- 4.Muto Y, Moriwaki H, Ninomiya M, Adachi S, Saito A, Takasaki K T, Tanaka T, Tsurumi K, Okuno M, Tomita E, Nakamura T, Kojima T. N Engl J Med. 1996;334:1561–1567. doi: 10.1056/NEJM199606133342402. [DOI] [PubMed] [Google Scholar]
- 5.Langenfeld J, Lonardo F, Kiyokawa H, Passalaris T, Ahn M-J, Rusch V, Dmitrovsky E. Oncogene. 1996;13:1983–1990. [PubMed] [Google Scholar]
- 6.Sherr C J. Cell. 1994;79:551–555. doi: 10.1016/0092-8674(94)90540-1. [DOI] [PubMed] [Google Scholar]
- 7.Nurse P. Cell. 1994;79:547–550. doi: 10.1016/0092-8674(94)90539-8. [DOI] [PubMed] [Google Scholar]
- 8.Sherr C J, Roberts J M. Genes Dev. 1995;9:1149–1163. doi: 10.1101/gad.9.10.1149. [DOI] [PubMed] [Google Scholar]
- 9.Nasmyth K. Science. 1996;274:1643–1645. doi: 10.1126/science.274.5293.1643. [DOI] [PubMed] [Google Scholar]
- 10.Resnitzky D, Gossen M, Bujard H, Reed S I. Mol Cell Biol. 1994;14:1669–1679. doi: 10.1128/mcb.14.3.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koff A, Cross F, Fisher A, Schumacher J, Leguellec K, Phillippe M, Roberts J M. Cell. 1991;66:1217–1228. doi: 10.1016/0092-8674(91)90044-y. [DOI] [PubMed] [Google Scholar]
- 12.Koff A, Giordano A, Desai D, Yamashita K, Harper J W, Elledge S, Nishimoto T, Morgan D O, Franza B R, Roberts J M. Science. 1992;257:1689–1694. doi: 10.1126/science.1388288. [DOI] [PubMed] [Google Scholar]
- 13.Ohtsubo M, Theodoras A M, Schumacher J, Roberts J M, Pagano M. Mol Cell Biol. 1995;15:2612–2624. doi: 10.1128/mcb.15.5.2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murray A. Cell. 1995;81:149–152. doi: 10.1016/0092-8674(95)90322-4. [DOI] [PubMed] [Google Scholar]
- 15.Hochstrasser M. Cell. 1996;84:813–815. doi: 10.1016/s0092-8674(00)81058-2. [DOI] [PubMed] [Google Scholar]
- 16.Klotzbucher A, Stewart E, Harrison D, Hunt T. EMBO J. 1996;15:3053–3064. [PMC free article] [PubMed] [Google Scholar]
- 17.Wilkinson K D. Annu Rev Nutr. 1995;15:161–189. doi: 10.1146/annurev.nu.15.070195.001113. [DOI] [PubMed] [Google Scholar]
- 18.Mahaffey D T, Yoo Y, Rechsteiner M. FEBS Lett. 1995;370:109–112. doi: 10.1016/0014-5793(95)00799-f. [DOI] [PubMed] [Google Scholar]
- 19.Aristarkhov A, Eytan E, Moghe A, Admon A, Hershko A, Ruderman J V. Proc Natl Acad Sci USA. 1996;93:4294–4299. doi: 10.1073/pnas.93.9.4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hershko A, Ganoth D, Sudakin V, Dahan A, Cohen L H, Luca F C, Ruderman J V, Eytan E. J Biol Chem. 1994;269:4940–4946. [PubMed] [Google Scholar]
- 21.Deshaies R J, Chau V, Kirschner M. EMBO J. 1995;14:303–312. doi: 10.1002/j.1460-2075.1995.tb07004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ciechanover A. Cell. 1994;79:13–21. doi: 10.1016/0092-8674(94)90396-4. [DOI] [PubMed] [Google Scholar]
- 23.Omura S, Matsuzaki K, Fujimoto T, Kosuge K, Furuya T, Fujita S, Nakagawa A. J Antibiot. 1991;44:117–118. doi: 10.7164/antibiotics.44.117. [DOI] [PubMed] [Google Scholar]
- 24.Reddel R R, Ke Y, Gerwin B I, McMenamin M G, Lechner J F, Su R T, Brash D E, Park J-B, Rhim J S, Harris C C. Cancer Res. 1988;48:1904–1909. [PubMed] [Google Scholar]
- 25.Lechner J F, Haugen A, McClendon I A, Pettis E W. In Vitro. 1982;18:633–642. doi: 10.1007/BF02796396. [DOI] [PubMed] [Google Scholar]
- 26.Ahn M-J, Langenfeld J, Moasser M M, Rusch V, Dmitrovsky E. Oncogene. 1995;11:2357–2364. [PubMed] [Google Scholar]
- 27.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. New York: Wiley; 1992. [Google Scholar]
- 28.Jackson R J, Hunt T. Methods Enzymol. 1983;96:50–74. doi: 10.1016/s0076-6879(83)96008-1. [DOI] [PubMed] [Google Scholar]
- 29.Hunt T, Vanderhoff G, London I M. J Mol Biol. 1972;66:471–481. doi: 10.1016/0022-2836(72)90427-5. [DOI] [PubMed] [Google Scholar]
- 30.Won K-A, Reed S I. EMBO J. 1996;15:4182–4193. [PMC free article] [PubMed] [Google Scholar]
- 31.Diehl J A, Zindy F, Sherr C J. Genes Dev. 1997;11:957–972. doi: 10.1101/gad.11.8.957. [DOI] [PubMed] [Google Scholar]
- 32.Rogers S, Wells R, Rechsteiner M. Science. 1986;234:364–369. doi: 10.1126/science.2876518. [DOI] [PubMed] [Google Scholar]
- 33.Yoshida H, Kitamura K, Tanaka K, Omura S, Miyazaki T, Hachiya T, Ohno R, Naoe T. Cancer Res. 1996;56:2945–2948. [PubMed] [Google Scholar]
- 34.Schauer I E, Siriwardana S, Langan T A, Sclafani R A. Proc Natl Acad Sci USA. 1994;91:7827–7831. doi: 10.1073/pnas.91.16.7827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Keyomarsi K, O’Leary N, Molnar G, Lees E, Fingert H J, Pardee A B. Cancer Res. 1994;54:380–385. [PubMed] [Google Scholar]
- 36.Dutta A, Chandra R, Leiter L M, Lester S. Proc Natl Acad Sci USA. 1995;92:5386–5390. doi: 10.1073/pnas.92.12.5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bortner D M, Rosenberg M P. Mol Cell Biol. 1997;17:453–459. doi: 10.1128/mcb.17.1.453. [DOI] [PMC free article] [PubMed] [Google Scholar]