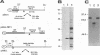Abstract
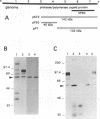
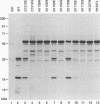
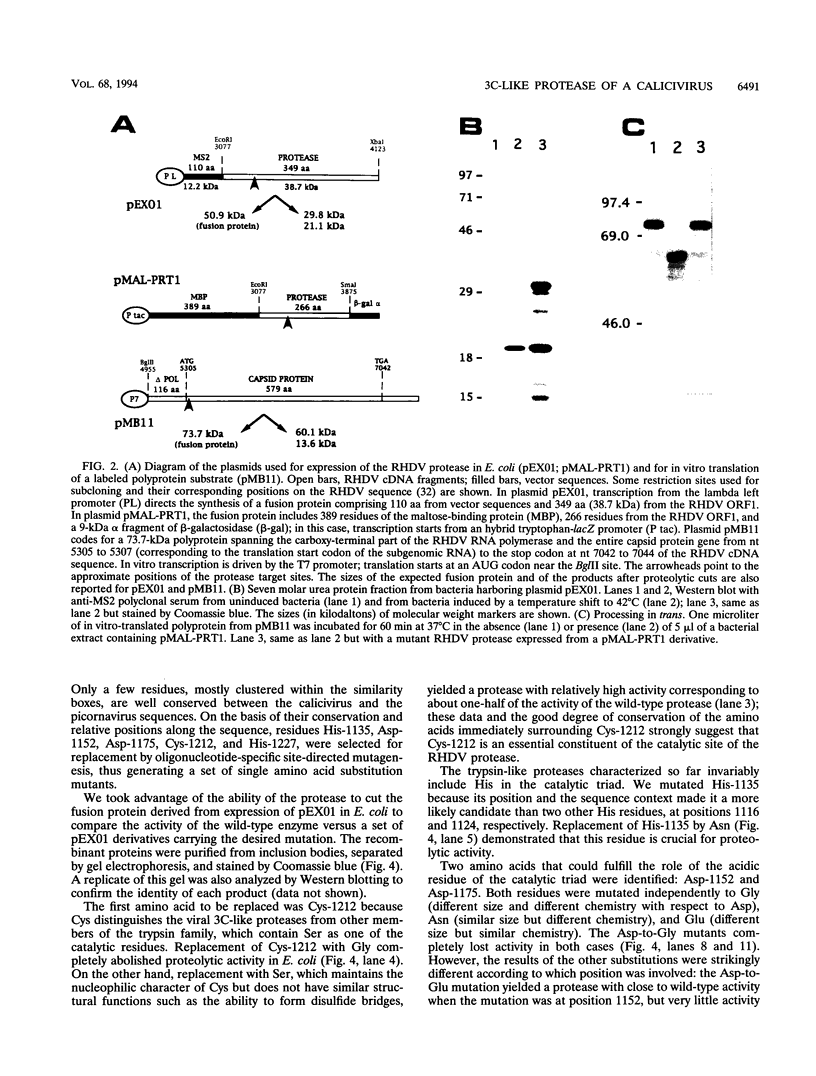
Expression studies conducted in vitro and in Escherichia coli led to the identification of a protease from rabbit hemorrhagic disease virus (RHDV). The gene coding for this protease was found to be located in the central part of the genome preceding the putative RNA polymerase gene. It was demonstrated that the protease specifically cuts RHDV polyprotein substrates both in cis and in trans. Site-directed mutagenesis experiments revealed that the RHDV protease closely resembles the 3C proteases of picornaviruses with respect to the amino acids directly involved in the catalytic activity as well as to the role played by histidine as part of the substrate binding pocket.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allaire M., Chernaia M. M., Malcolm B. A., James M. N. Picornaviral 3C cysteine proteinases have a fold similar to chymotrypsin-like serine proteinases. Nature. 1994 May 5;369(6475):72–76. doi: 10.1038/369072a0. [DOI] [PubMed] [Google Scholar]
- Baum E. Z., Bebernitz G. A., Palant O., Mueller T., Plotch S. J. Purification, properties, and mutagenesis of poliovirus 3C protease. Virology. 1991 Nov;185(1):140–150. doi: 10.1016/0042-6822(91)90762-z. [DOI] [PubMed] [Google Scholar]
- Bazan J. F., Fletterick R. J. Viral cysteine proteases are homologous to the trypsin-like family of serine proteases: structural and functional implications. Proc Natl Acad Sci U S A. 1988 Nov;85(21):7872–7876. doi: 10.1073/pnas.85.21.7872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blok V. C., Wardell J., Jolly C. A., Manoukian A., Robinson D. J., Edwards M. L., Mayo M. A. The nucleotide sequence of RNA-2 of raspberry ringspot nepovirus. J Gen Virol. 1992 Sep;73(Pt 9):2189–2194. doi: 10.1099/0022-1317-73-9-2189. [DOI] [PubMed] [Google Scholar]
- Carter M. J., Milton I. D., Meanger J., Bennett M., Gaskell R. M., Turner P. C. The complete nucleotide sequence of a feline calicivirus. Virology. 1992 Sep;190(1):443–448. doi: 10.1016/0042-6822(92)91231-i. [DOI] [PubMed] [Google Scholar]
- Carter M. J., Milton I. D., Turner P. C., Meanger J., Bennett M., Gaskell R. M. Identification and sequence determination of the capsid protein gene of feline calicivirus. Arch Virol. 1992;122(3-4):223–235. doi: 10.1007/BF01317185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheah K. C., Leong L. E., Porter A. G. Site-directed mutagenesis suggests close functional relationship between a human rhinovirus 3C cysteine protease and cellular trypsin-like serine proteases. J Biol Chem. 1990 May 5;265(13):7180–7187. [PubMed] [Google Scholar]
- Dessens J. T., Lomonossoff G. P. Mutational analysis of the putative catalytic triad of the cowpea mosaic virus 24K protease. Virology. 1991 Oct;184(2):738–746. doi: 10.1016/0042-6822(91)90444-g. [DOI] [PubMed] [Google Scholar]
- Dougherty W. G., Parks T. D., Cary S. M., Bazan J. F., Fletterick R. J. Characterization of the catalytic residues of the tobacco etch virus 49-kDa proteinase. Virology. 1989 Sep;172(1):302–310. doi: 10.1016/0042-6822(89)90132-3. [DOI] [PubMed] [Google Scholar]
- Forss S., Strebel K., Beck E., Schaller H. Nucleotide sequence and genome organization of foot-and-mouth disease virus. Nucleic Acids Res. 1984 Aug 24;12(16):6587–6601. doi: 10.1093/nar/12.16.6587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A. E., Donchenko A. P., Blinov V. M., Koonin E. V. Cysteine proteases of positive strand RNA viruses and chymotrypsin-like serine proteases. A distinct protein superfamily with a common structural fold. FEBS Lett. 1989 Jan 30;243(2):103–114. doi: 10.1016/0014-5793(89)80109-7. [DOI] [PubMed] [Google Scholar]
- Harris K. S., Reddigari S. R., Nicklin M. J., Hämmerle T., Wimmer E. Purification and characterization of poliovirus polypeptide 3CD, a proteinase and a precursor for RNA polymerase. J Virol. 1992 Dec;66(12):7481–7489. doi: 10.1128/jvi.66.12.7481-7489.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellen C. U., Fäcke M., Kräusslich H. G., Lee C. K., Wimmer E. Characterization of poliovirus 2A proteinase by mutational analysis: residues required for autocatalytic activity are essential for induction of cleavage of eukaryotic initiation factor 4F polypeptide p220. J Virol. 1991 Aug;65(8):4226–4231. doi: 10.1128/jvi.65.8.4226-4231.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hämmerle T., Hellen C. U., Wimmer E. Site-directed mutagenesis of the putative catalytic triad of poliovirus 3C proteinase. J Biol Chem. 1991 Mar 25;266(9):5412–5416. [PubMed] [Google Scholar]
- Ivanoff L. A., Towatari T., Ray J., Korant B. D., Petteway S. R., Jr Expression and site-specific mutagenesis of the poliovirus 3C protease in Escherichia coli. Proc Natl Acad Sci U S A. 1986 Aug;83(15):5392–5396. doi: 10.1073/pnas.83.15.5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X., Wang M., Wang K., Estes M. K. Sequence and genomic organization of Norwalk virus. Virology. 1993 Jul;195(1):51–61. doi: 10.1006/viro.1993.1345. [DOI] [PubMed] [Google Scholar]
- Jore J., De Geus B., Jackson R. J., Pouwels P. H., Enger-Valk B. E. Poliovirus protein 3CD is the active protease for processing of the precursor protein P1 in vitro. J Gen Virol. 1988 Jul;69(Pt 7):1627–1636. doi: 10.1099/0022-1317-69-7-1627. [DOI] [PubMed] [Google Scholar]
- Kean K. M., Teterina N. L., Marc D., Girard M. Analysis of putative active site residues of the poliovirus 3C protease. Virology. 1991 Apr;181(2):609–619. doi: 10.1016/0042-6822(91)90894-h. [DOI] [PubMed] [Google Scholar]
- Kowalski H., Maurer-Fogy I., Zorn M., Mischak H., Kuechler E., Blaas D. Cleavage site between VP1 and P2A of human rhinovirus is different in serotypes 2 and 14. J Gen Virol. 1987 Dec;68(Pt 12):3197–3200. doi: 10.1099/0022-1317-68-12-3197. [DOI] [PubMed] [Google Scholar]
- Kräusslich H. G., Wimmer E. Viral proteinases. Annu Rev Biochem. 1988;57:701–754. doi: 10.1146/annurev.bi.57.070188.003413. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- La Monica N., Meriam C., Racaniello V. R. Mapping of sequences required for mouse neurovirulence of poliovirus type 2 Lansing. J Virol. 1986 Feb;57(2):515–525. doi: 10.1128/jvi.57.2.515-525.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lambden P. R., Caul E. O., Ashley C. R., Clarke I. N. Sequence and genome organization of a human small round-structured (Norwalk-like) virus. Science. 1993 Jan 22;259(5094):516–519. doi: 10.1126/science.8380940. [DOI] [PubMed] [Google Scholar]
- Lawson M. A., Semler B. L. Poliovirus thiol proteinase 3C can utilize a serine nucleophile within the putative catalytic triad. Proc Natl Acad Sci U S A. 1991 Nov 15;88(22):9919–9923. doi: 10.1073/pnas.88.22.9919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margis R., Pinck L. Effects of site-directed mutagenesis on the presumed catalytic triad and substrate-binding pocket of grapevine fanleaf nepovirus 24-kDa proteinase. Virology. 1992 Oct;190(2):884–888. doi: 10.1016/0042-6822(92)90931-e. [DOI] [PubMed] [Google Scholar]
- Margis R., Ritzenthaler C., Reinbolt J., Pinck M., Pinck L. Genome organization of grapevine fanleaf nepovirus RNA2 deduced from the 122K polyprotein P2 in vitro cleavage products. J Gen Virol. 1993 Sep;74(Pt 9):1919–1926. doi: 10.1099/0022-1317-74-9-1919. [DOI] [PubMed] [Google Scholar]
- Meyers G., Wirblich C., Thiel H. J. Genomic and subgenomic RNAs of rabbit hemorrhagic disease virus are both protein-linked and packaged into particles. Virology. 1991 Oct;184(2):677–686. doi: 10.1016/0042-6822(91)90437-G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers G., Wirblich C., Thiel H. J. Rabbit hemorrhagic disease virus--molecular cloning and nucleotide sequencing of a calicivirus genome. Virology. 1991 Oct;184(2):664–676. doi: 10.1016/0042-6822(91)90436-f. [DOI] [PubMed] [Google Scholar]
- Neill J. D. Nucleotide sequence of a region of the feline calicivirus genome which encodes picornavirus-like RNA-dependent RNA polymerase, cysteine protease and 2C polypeptides. Virus Res. 1990 Nov;17(3):145–160. doi: 10.1016/0168-1702(90)90061-f. [DOI] [PubMed] [Google Scholar]
- Parks G. D., Baker J. C., Palmenberg A. C. Proteolytic cleavage of encephalomyocarditis virus capsid region substrates by precursors to the 3C enzyme. J Virol. 1989 Mar;63(3):1054–1058. doi: 10.1128/jvi.63.3.1054-1058.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra F., Boga J. A., Marin M. S., Casais R. The amino terminal sequence of VP60 from rabbit hemorrhagic disease virus supports its putative subgenomic origin. Virus Res. 1993 Mar;27(3):219–228. doi: 10.1016/0168-1702(93)90034-k. [DOI] [PubMed] [Google Scholar]
- Parra F., Prieto M. Purification and characterization of a calicivirus as the causative agent of a lethal hemorrhagic disease in rabbits. J Virol. 1990 Aug;64(8):4013–4015. doi: 10.1128/jvi.64.8.4013-4015.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommergruber W., Zorn M., Blaas D., Fessl F., Volkmann P., Maurer-Fogy I., Pallai P., Merluzzi V., Matteo M., Skern T. Polypeptide 2A of human rhinovirus type 2: identification as a protease and characterization by mutational analysis. Virology. 1989 Mar;169(1):68–77. doi: 10.1016/0042-6822(89)90042-1. [DOI] [PubMed] [Google Scholar]
- Stanway G., Hughes P. J., Mountford R. C., Minor P. D., Almond J. W. The complete nucleotide sequence of a common cold virus: human rhinovirus 14. Nucleic Acids Res. 1984 Oct 25;12(20):7859–7875. doi: 10.1093/nar/12.20.7859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strebel K., Beck E., Strohmaier K., Schaller H. Characterization of foot-and-mouth disease virus gene products with antisera against bacterially synthesized fusion proteins. J Virol. 1986 Mar;57(3):983–991. doi: 10.1128/jvi.57.3.983-991.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoda H., Nicklin M. J., Murray M. G., Anderson C. W., Dunn J. J., Studier F. W., Wimmer E. A second virus-encoded proteinase involved in proteolytic processing of poliovirus polyprotein. Cell. 1986 Jun 6;45(5):761–770. doi: 10.1016/0092-8674(86)90790-7. [DOI] [PubMed] [Google Scholar]
- Ypma-Wong M. F., Dewalt P. G., Johnson V. H., Lamb J. G., Semler B. L. Protein 3CD is the major poliovirus proteinase responsible for cleavage of the P1 capsid precursor. Virology. 1988 Sep;166(1):265–270. doi: 10.1016/0042-6822(88)90172-9. [DOI] [PubMed] [Google Scholar]
- Yu S. F., Lloyd R. E. Identification of essential amino acid residues in the functional activity of poliovirus 2A protease. Virology. 1991 Jun;182(2):615–625. doi: 10.1016/0042-6822(91)90602-8. [DOI] [PubMed] [Google Scholar]