Abstract
Hox complex genes control spatial patterning mechanisms in the development of arthropod and vertebrate body plans. Hox genes are all expressed during embryogenesis in these groups, which are all directly developing organisms in that embryogenesis leads at once to formation of major elements of the respective adult body plans. In the maximally indirect development of a large variety of invertebrates, the process of embryogenesis leads only to a free-living, bilaterally organized feeding larva. Maximal indirect development is exemplified in sea urchins. The 5-fold radially symmetric adult body plan of the sea urchin is generated long after embryogenesis is complete, by a separate process occurring within imaginal tissues set aside in the larva. The single Hox gene complex of Strongylocentrotus purpuratus contains 10 genes, and expression of eight of these genes was measured by quantitative methods during both embryonic and larval developmental stages and also in adult tissues. Only two of these genes are used significantly during the entire process of embryogenesis per se, although all are copiously expressed during the stages when the adult body plan is forming in the imaginal rudiment. They are also all expressed in various combinations in adult tissues. Thus, development of a microscopic, free-living organism of bilaterian grade, the larva, does not appear to require expression of the Hox gene cluster as such, whereas development of the adult body plan does. These observations reflect on mechanisms by which bilaterian metazoans might have arisen in Precambrian evolution.
The Hox gene cluster occupies a central position in current conceptions of both the development and evolution of metazoan body plans. Yet systematic evidence regarding the developmental expression of these genes is largely confined to two animal groups, the arthropods and the chordates. These groups are both direct developers, in the sense that major aspects of their adult body plans form immediately during embryogenesis, e.g., the head and the anterior/posterior body axis, the major mesodermal anlagen, the central nervous system, and metameric body structures (1). Two other organisms for which some information about developmental Hox gene expression exists viz., Caenorhabditis elegans (2) and leech (3, 4), are also direct developers. Expression of the Hox complex has never been examined systematically in any animal that displays maximal indirect development. Here the process of embryogenesis produces a free-living, motile larva capable of feeding and growth, but in structure this larva bears essentially no resemblance to the adult body plan of the species. In maximal indirect development, the adult body plan instead forms within the larva by a complex secondary process from special patches of cells set aside during embryogenesis (1). The larva itself is a small, bilaterally organized metazoan organism that includes mesodermal as well as ectodermal, and endodermal cell types. Thus, it has muscle cells, neurons, gut cells, skeletogenic cells, and sensory and epidermal cells, some specialized with respect to their ciliary appurtenances, and it is equipped with a complete digestive tract including mouth, esophagus, stomach, intestine, and anus. Maximal indirect development affords the opportunity of a complete temporal separation between the embryonic process by which the larval micrometazoan develops, and the postembryonic process in which the adult body plan is organized.
In a common mode of embryonic specification, which appears primitive for bilaterian metazoans (5–7), the egg is divided up into blastomeres of more or less invariant lineage. Specification of given lineage elements depends on short-range interblastomere signaling occurring during cleavage, as well as on inherited consequences of the cytoarchitecture of the egg. The specification process directly generates a mosaic of blastomeres before any migratory cells appear, and the immediate progeny of these blastomeres give rise directly to differentiated cell types (type 1 embryonic process) (6, 7). Early development in most modern bilaterian clades operates in this way. Two exceptions are the highly derived syncytial strategy used in most insects and the almost unique processes that have evolved in vertebrate (but not invertebrate) chordates, wherein the large eggs divide to produce thousands of cells before transcriptional activation of the genome. In vertebrates, specification of cells in most regions of the embryo occurs without respect to lineage in large, migrating cell populations.
In all direct developing bilaterians, the processes of adult body plan formation are telescoped down upon the embryonic process, and basic components of the adult body plan emerge soon after gastrulation. However, in indirectly developing deuterostomes and lophotrochozoan (8) protostomes (i.e., polychaete annelids, some molluscs, flatworms, and brachiopods), we can see the nature of the product that the type 1 embryonic process is capable of generating on its own, i.e., in the absence of further growth, and of the more complex processes by which adult body plans are formed. This product is a small metazoan organism of simple construction, i.e., the larval form of each such species of animal. Because the embryonic blastomeres have a fixed division potential (other than the set-aside cells that are reserved for postembryonic development), these micrometazoan organisms consist of only a few thousand cells. We argued (5) in brief (i) that the ancestors of modern bilaterian metazoans developed by type 1 embryonic processes because this mode of early embryonic specification is a property shared by most extant bilaterian groups; (ii) that the regulatory apparatus underlying this mode of embryogenesis would have sufficed for the generation of a micrometazoan fauna, similar in grade of organization to the larvae of modern maximal indirect developers; (iii) that such a fauna provided the preexistent platform for evolution of the modern bilaterian clades; and (iv) that additional and much more complex developmental regulatory hardwiring would have been required for the advent of large animals displaying body plans of the complexity of modern bilaterians and all fossil forms recognized as such. Because the bilaterians are monophyletic, this augmentation in developmental regulatory capacity probably happened before divergence of the major bilaterian clades. Two fundamental and interrelated changes were proposed: the appearance of cell populations set aside from embryonic specification processes that retained indefinite division capacity and the erection of genetic regulatory apparatus for the patterning of these cell populations. The nature of such apparatus is now becoming apparent. Spatial patterning of body parts in the development of modern bilaterians is a stepwise process (refs. 5 and 9; see examples in insect imaginal disc patterning, ref. 10; and in limb bud development, ref. 11; brain development, ref. 12; and dorsal axial specification, ref. 13). A succession of regulatory states is set up by means of expression of genes dedicated to the patterning process that encodes transcription factors, in distinct spatial domains that foreshadow parts of the structure. These domains are organized developmentally by spatially confined signaling systems that operate upstream and downstream of the regulatory patterning genes. The Hox cluster genes operate within this system and play key roles in many aspects of spatial patterning (14, 15). We are beginning to understand how these affect morphological outcome by controlling other patterning functions (16, 17).
The sea urchin provides an excellent test case for a specific prediction deriving from these concepts. This prediction (5) was that the embryonic regulatory mechanisms needed to generate an organism of the relatively low complexity of the larva will exclude the use of regional patterning devices such as the Hox gene cluster, but, on the other hand, these genes must be called into action in the separate process of adult body plan formation. The feeding sea urchin larva bears virtually no relation to the 5-fold radially symmetrical adult echinoderm body plan that will develop within it. It is bilaterally and not radially organized, and neither the larval mouth nor its anus, its skeletal structures, its body wall, or its neuronal components become equivalent components of the juvenile. Nor are the anterior-posterior or dorsoventral axes of the larva preserved in the adult body plan. There were of course already some indications from direct developing animals that Hox cluster genes do not control specification processes in type 1 embryogenesis. For example, in C. elegans an almost normal embryo and first-stage larva are formed irrespective of mutation of the genes of the reduced Hox gene cluster of this organism (18). In the leech the Hox genes are activated only after the formation of the segmented body plan has occurred by elaboration of the germ band, the cells of which are previously segmentally specified, at their birth (19).
The complete Hox gene cluster of Strongylocentrotus purpuratus, a typical indirectly developing sea urchin, has now been cloned and mapped (P.M., J. P. Rast, C.A.-M., and E.H.D., unpublished data), and 10 genes, which according to their sequence have been assigned to the equivalent Hox paralogue groups, have been identified within it. This work provided the gene-specific probes that were used in the following experiments.
MATERIALS AND METHODS
Probe excess RNA titrations were performed as described (20), except for the following details. Increasing amounts of total RNA from each developmental stage and sufficient yeast tRNA to a total of 60 μg were coprecipitated in ethanol with 0.1 ng of purified riboprobe in the presence of 0.2 M ammonium acetate. The precipitated RNA was pelleted, washed with ethanol, and lyophilized. The hybridization was performed in 20 μl of 50% formamide, 0.4 M NaCl, 1 mM EDTA, and 25 mM Pipes (pH 7) at 50°C for 20 h. Unhybridized RNA was removed by digestion with 500 units/ml T1 RNase and 40 μg/ml RNase A at 37°C for 30 min in 0.15 M NaCl, 30 mM Tris (pH 8), and 2 mM EDTA. RNA duplexes were precipitated in the presence of 100 μg of yeast RNA by the addition of 1 volume of 10% trichloroacetic acid. The precipitated RNA was collected on glass fiber filters, washed, and dried, and the amount of radioactive RNA was determined by scintillation counting.
Whole-mount in situ hybridization (WMISH) was performed by the method of Holland et al. (21) with the following modifications: larvae raised in the laboratory (22) were fixed overnight at 0°C in 3.2% formaldehyde and 0.2 M phosphate buffer, pH 7.4, with total [Na+] brought to 0.55 M by addition of NaCl. Hybridization was in the same phosphate-NaCl buffer, containing 65% formamide and 0.1% Tween 20, at 50°C for 5 h. For the alkaline phosphatase reaction N,N-dimethylformamide was added to the buffer to 10% to increase sensitivity.
RESULTS
Earlier work had shown that two of the S. purpuratus Hox genes are expressed during embryogenesis in particular structures. When the Hox cluster was resolved (P.M., J. P. Rast, C.A.-M., and E.H.D., unpublished data), these two genes were seen to be a paralogue of Hox7 (SpHox7, original “Hbox1,” ref. 23) and a paralogue of Hox11/13 (SpHox11/13b, originally “Hbox7,” ref. 24). The S. purpuratus cluster contains 10 genes but differs slightly from the vertebrate consensus in that it has only one gene of the Hox4 and 5 type (SpHox4/5). Of the three posterior genes, SpHox9/10 is more closely related to Hox9 and Hox10 than to Hox11–13, whereas the others, SpHox11/13a and SpHox11/13b, are more similar to the latter (P.M., J. P. Rast, C.A.-M., and E.H.D., unpublished data). The SpHox7 and SpHox11/13b genes had been found to be activated before gastrulation, and by late embryogenesis their products are confined, respectively, to the vertex region of the aboral ectoderm (23) and to subregions of the oral ectoderm, the larval arms, and the foregut (24). There was little significant evidence regarding utilization of the other Hox genes in development. Because negative WMISH data are not informative, we decided to measure the number of transcripts of each of eight different Hox cluster genes in the RNA of embryos collected at different stages: in larvae during the period when the adult body plan is developing and in adult tissues. Transcript numbers were calculated from probe excess titrations by using antisense RNA probes specific for each Hox gene. For SpHox7 and SpHox11/13b, these probes were obtained from the respective embryo cDNAs, and for the other genes of the cluster, the probes were obtained from genomic subclones. The probe excess titration method (20) offered several significant advantages for these purposes: (i) it is extremely sensitive, permitting reliable detection of <0.05 mRNA molecules per average embryonic cell; (ii) it is impervious to any but very severe RNA degradation, because the probe is present in excess, and the unit size of the protected product is not required for detection; (iii) hybridization is kinetically independent of the level of expression, because the reactions are uniformly driven to completion in efficient solution hybridization reactions, carried to high R0t with respect to the probe (22). As controls for the measurement procedures and calculations, titrations were carried out for mRNAs of the transcription factor gene SpZ12–1 (25) and of the cytoskeletal actin CyIIIa (26, 27), the same RNAs as used for estimation of Hox gene transcripts. The values observed were close to those previously published.
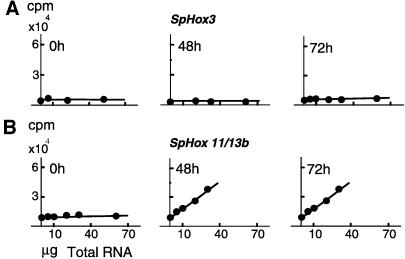
Representative titration data are shown for SpHox3 and SpHox7 transcripts in Fig. 1. Transcript numbers per unit mass RNA and per embryo are directly proportional to the absolute slopes of each data set. In this example, SpHox3 was not expressed significantly at any embryonic stage; the slopes were close to 0 until late in embryogenesis, and at 72 h, the number of transcripts represented by the low positive slope indicates only about 100 transcripts in the whole embryo (or ≈0.06 molecule per average cell). An embryo of this age contains 1,800 cells and is able to feed, because it is equipped with a complete digestive tract (Fig. 2A). SpHox3 transcripts are abundant in the 2-week larva, however; i.e., there are ≈10,000 molecules per organism as calculated from this data set. At this stage, adult body plan formation has begun. In contrast to the pattern of expression shown in Fig. 1A, in Fig. 1B the data show that the SpHox7 gene was expressed at a relatively high level during embryogenesis (these measurements are in satisfactory agreement with those published earlier, ref. 23); there were>6,000 transcripts per embryo at all stages from blastula on (for values see, Fig. 3). SpHox7 is also used in the larval stages because the number of transcripts had increased further, by about 2-fold. Correlation coefficients for these titration curves were >95% and in general were of this magnitude or better for all the measurements summarized in the following.
Figure 1.
Single-strand probe excess titrations of transcripts from two S. purpuratus Hox genes: SpHox3 (A) and SpHox11/13b (B). These examples are representative of the data sets that provided the measurements reported in this paper. Total RNA was extracted from unfertilized eggs and embryos at the indicated stages and in increasing amounts reacted with 32P-labeled antisense RNA probes (see legend to Fig. 3 and Materials and Methods for probes and procedures). Transcript numbers were calculated from the slopes as described (20), taking into account the amount of total RNA per embryo (2.8 ng; ref. 22) and the lengths and specific activities of the probes.
Figure 2.
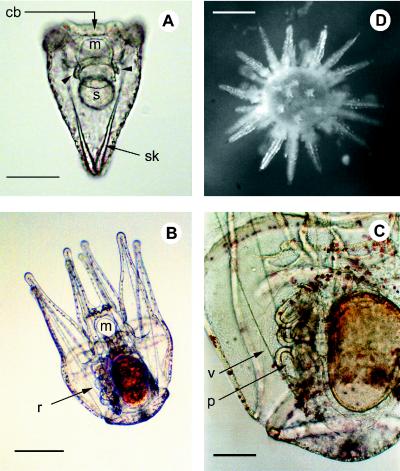
S. purpuratus developmental stages. (A) Pluteus stage, termination of embryogenesis (96 h; essentially the same as the 72-h stage at which the measurements of Fig. 3 were made). m, Mouth; s, stomach; sk, skeleton; cb, ciliated band; coelomic sacs (arrowhead). (Bar = 50 μm.) (B) Larva 3 weeks after feeding begins. (Bar = 250 μm.) (C) Close-up view of rudiment in a larvae at the same stage. r, Rudiment; v, vestibule; p, primary podia. (Bar = 100 μm.) (D) Newly emergent juvenile sea urchin. (Bar = 400 μm.)
Figure 3.
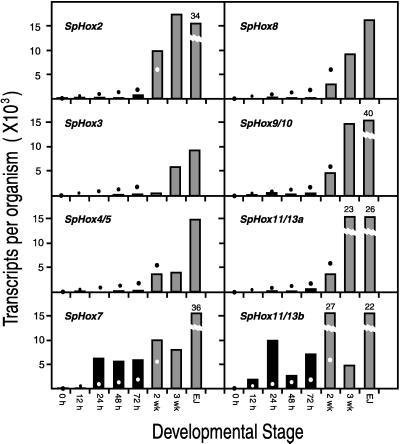
Transcripts of Hox cluster genes per embryo, larva, or newly emergent juvenile. Histograms indicate transcript number for the indicated stages of embryogenesis: 0 h, unfertilized egg; 12 h, late cleavage; 24 h, mesenchyme blastula; 48 h, late gastrula; 72 h, completed larva (see Fig. 2A); 2-wk, larva 2 weeks after fertilization; 3-wk, larva at 3 weeks of development (Fig. 2 B and C); EJ, emergent juvenile, a few days after metamorphosis. Broken bars indicate values at top rather than ordinate values. The dots indicate the number of cells per organism (same ordinates). All embryonic stages have about 2.8 ng of total RNA (23); 2-wk larvae have about 22.5 ng (27); 3-wk larvae have about 35 ng (27); emergent juveniles have about 130 ng (C.A.-M. and E.H.D. data). Probes were as follows: SpHox2, 167-bp exon sequence extending into an intron and including 106 bp from the 5′ end of the homeobox; SpHox3, 310 bp of exon sequence, including 111 bp from the 3′ end of the homeobox; SpHox4/5, 589 bp exon sequence, partially in 3′-untranslated region and including 105 bp of 3′ homeobox region; SpHox7, 595 bp insert from a S. purpuratus cDNA clone; SpHox8, 493-bp exon sequence extending into 3′-untranslated region; SpHox9/10, 227-bp exon sequence extending into 3′-untranslated region (a kind gift of R. Maxson); SpHox11/13a, 555-bp exon sequence extending into 3′-untranslated region; SpHox11/13b, 562-bp subclone from a cDNA clone (provided by R. Maxson). Reproducibility in transcript numbers was generally within ±30%, except when transcript representation was near the lower limit of detection (<100 molecules per embryo) when variation was about 2-fold around the mean. For example, measurements with two entirely different 72-h embryo RNA preparations gave about 70 and 200 molecules for SpHox8 and 5,500 and 6,200 molecules for SpHox7.
Expression of the S. purpuratus Hox Gene Complex in Embryos and Larvae.
Results for all eight Hox genes studied are given in Fig. 3. The ordinates show the number of transcripts per embryo for each Hox gene at the indicated stages of embryogenesis (left columns) and per larva or newly emergent juvenile (right columns). The larvae were harvested at 2 weeks and 3 weeks (Fig. 2 B and C), when the rudiment has advanced to the point where the five primary podia and tooth sacs are already beginning to protrude from the ventral surface (for a brief recent review of early rudiment development, see ref. 28; for detailed account, ref. 29). Metamorphosis occurred at about 5 to 6 weeks in the laboratory. Emergent juveniles (Fig. 2D) were harvested shortly thereafter. The dots in Fig. 3 indicate the total number of cells at each stage measured, read off the same ordinates, and thus the number of transcripts required for a single molecule per average cell. During embryological development, from egg to 72-h pluteus stage, the mass of total RNA and of mRNA remains essentially constant (22), but after feeding begins there is an enormous increase in cell constituents. By metamorphosis, the larva contains about 1.5 × 105 cells, >90% of which are in the imaginal rudiment and other newly formed tissue elements that will contribute to the structures of the juvenile (26). In most cases, only a single data set is included for each probe in Fig. 3, and the measurements shown in each panel were carried out at the same time on the whole set of RNA preparations with the same probe preparation. Very similar results were obtained in replicate titrations carried out with different RNA preparations or with different probe preparations by using the same RNA preparation.
The most striking result in Fig. 3 is that SpHox2, SpHox3, SpHox4/5, SpHox8, SpHox9/10, and SpHox11/13a are not expressed significantly at any stage of embryogenesis. mRNAs from these genes are either not detectable at all or just above the limits of detection of the very sensitive titration method (cf. Fig. 1A). Fig. 3 shows that only SpHox7 and SpHox11/13b, as previously shown (23, 24), are expressed significantly in the embryo.
By 3 weeks fertilization has begun and the picture has changed completely. Fig. 3 shows that transcripts of all of the eight Hox cluster genes studied are present at significant levels, and this is true for most of the genes seen at the 2-week early rudiment stage (28). The accumulation of transcripts undoubtedly indicates Hox gene expression, and the suggestion that this expression is regional is supported by those WMISH observations of larval stages so far available. An example is shown in Fig. 4 for SpHox3. This gene was expressed in five patches of cells in the ventral region of the rudiment, apparently in the somatocoelar mesoderm of the nascent tooth sacs. From these structures derive not only the oral masticatory apparatus but also the hyponeural sinuses beneath which lie the radially organized central nervous system (29). SpHox3 was expressed specifically in the 5-fold radially symmetric organization of the adult body plan. There was no expression detectable by WMISH in larval as opposed to rudiment tissues or in the late embryo, excluding for this gene the possibility of even a few cells active at the levels of SpHox7 or SpHox11/13b.
Figure 4.
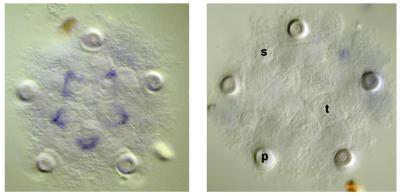
SpHox3 expression in the imaginal rudiment of a 3 week-old larva. (Left) Transcripts were localized by WMISH. (Right) A control WMISH localized by using a sense probe is also shown. p, Primary podia; s, adult spines; t, tooth sacs.
In summary, Fig. 3 shows that the genes for the Hox cluster are all represented by transcripts at levels ranging from a few thousand to more than 20,000 molecules per larva during the stages of rudiment formation. Levels in newly emergent juveniles are in every case even higher. In sea urchins, development of the adult form continues after metamorphosis; for example, the oral apparatus, mouth, esophagus, and hindgut are all yet to form at the time of emergence. Thus, Hox gene expression occurs throughout the period of adult body plan formation.
Expression in Adult Tissues.
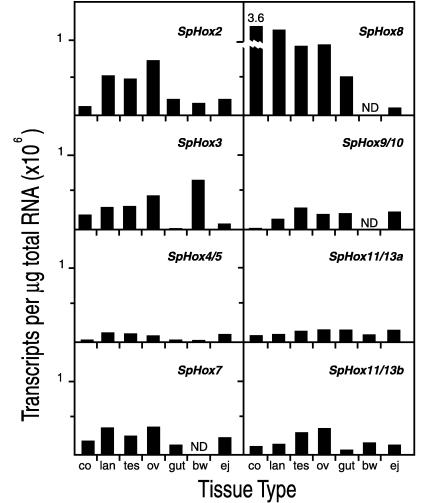
The measurements in Fig. 5 give transcript numbers per unit mass of total RNA (μg) for adult sea urchin coelomocytes: lantern apparatus (including both muscle and other connective tissue and portions of the central nervous system), testis, ovary, gut (stomach plus intestine), and body wall (epidermis plus spine musculature). Each of the eight genes is expressed in a certain pattern in these adult tissues, and conversely each tissue expresses a certain pattern of Hox complex genes. For example, coelomocytes, which constitute the immune system of sea urchins (30), expressed SpHox8 far more than any other of the genes, whereas they hardly expressed SpHox2, 4/5, 9/10, 11/13a, or 11/13b. Ovary and testis express SpHox2, 3, 7, 8, 9/10, and 11/13b, but SpHox4/5 and SpHox11/13a are expressed much less. Body wall expressed SpHox3 preferentially. Furthermore there was no obvious difference in the patterns of expression in adult tissues of these genes according to their positions in the cluster. For example, the pattern of expression of SpHox2, the most “anterior” of the genes studied, is most similar to that of SpHox11/13b, the most “posterior” gene in the cluster (P.M., J. P. Rast, C.A.-M., and E.H.D., unpublished data). Expression in the adult tissues would appear to indicate an other than developmental role in cell types, such as the coelomocytes, or organs, such as gut or lantern apparatus, although there could be continuing differentiation of certain cell populations occurring in each. It may be significant for the proposition that the Hox genes are supporting particular states of differentiation in adult sea urchins that the most clearly defined cell population here, the coelomocytes [there are five coelomocyte cell types (30), displays the sharpest preference profile, i.e., for SpHox8.
Figure 5.
Transcripts of Hox cluster genes in adult tissues. The presentation is as in Fig. 3, except that numbers of transcripts are given per μg of total RNA. co, Coelomocyte; lan, lantern; tes, testis; ov, ovary; bw, body wall; ej, emergent juvenile.
DISCUSSION
Hox Genes and Development of the Sea Urchin Embryo/Larva.
Fig. 3 shows that all of the tested genes of the Hox cluster function during the period of rudiment development, but much additional work will be required before we understand their roles. A particularly fascinating issue is the relation between the radially symmetrical echinoderm body plan and the “anterior-posterior” polarity of the Hox gene cluster. Detailed studies of the expression patterns of these genes within the growing rudiment, now in progress, will hopefully illuminate this issue. Whatever their role in adult body plan formation, our evidence indicates that, except for SpHox7 and SpHox11/13b, the Hox genes we studied play no significant role in the formation of the embryo/larva, i.e., in the development of larval structures, exclusive of the set-aside cell progeny that contribute to the imaginal rudiment from which the juvenile sea urchin derives. It is not even likely that SpHox7 or SpHox11/13b participate in regional embryonic specification functions. SpHox7 transcripts are first detected at the blastula stage throughout the region occupied by aboral ectoderm precursors (23). However, the aboral ectoderm is specified much earlier in development, and except for some boundary areas where the ciliated band forms later, the cells of which it is composed are fully committed by late blastula. Thus, all cells of the aboral ectoderm lineage have long before initiated expression of terminal aboral ectoderm-specific genes (28). Late in embryonic development, SpHox7 transcripts become confined to the vertex of the aboral ectoderm (23), which is constituted by the lineage of only one of the aboral ectoderm founder cells (31). During postembryonic life, skeletal elements later incorporated into the rudiment begin to form within this region, and perhaps SpHox7 expression carries out a function related to that process. SpHox11/13b expression is also activated at the blastula stage but is unlikely to play a role in regional specification because its transcripts are then distributed ubiquitously, as is also the protein (24). The RNA and protein are later found in multiple regions of the embryo, which are related neither in morphogenetic function nor by lineage, including the ciliated band, the tip of the archenteron, the oral ectoderm, and some regions of the forming larval arms (24). These areas are all sites where cell division continues or resumes late in embryonic development, and SpHox11/13b could be engaged in cell division control. This protein is in fact capable of binding to a target site in the cis-regulatory activator of a late histone gene (24, 32).
As discussed, the small number of transcripts of the other six Hox cluster genes found in the embryo before feeding do not appear to indicate expression that is of functional significance for the processes of embryogenesis. Throughout embryogenesis the levels of transcripts are either altogether undetectable or there are only a few hundred transcripts per organism. In the 48-h 1,000 cell embryo, the numbers of transcripts are, for SpHox2, ≈330 per embryo; for SpHox3, undetectable; for SpHox4/5, ≈250; for SpHox6, ≈150, for SpHox9/10, ≈190; for SpHox11/13a, ≈130; and the same or lower values obtain for earlier embryos. However, regional specification processes in this embryo are essentially complete by 48 h, although further elaboration of some differentiated cells and further morphogenetic progression takes place in archenteron, skeleton, ciliated band, and stomodaeum. On the other hand, the numbers of transcripts of SpHox7 and SpHox11/13b are quite respectable for genes encoding transcription factors. Thus, there are 30–100 SpHox7 transcripts per active cell between 48 and 72 h (23), and for SpHox11/13b, there are ≥30–40 transcripts per active cell (Fig. 3 and ref. 24). Were the other six genes to display transcripts at these levels, they would be confined to about two to eight cells in the late embryo. Although such a possibility cannot be excluded for most of these genes, the structures that constitute the late embryo, e.g., gut, oral ectoderm, skeletal structures, are all composed of much larger populations of cells. Furthermore, where we looked by WMISH, for instance by using SpHox3 probes, no actively expressing cells can, in fact, be observed at any embryonic stage. It is not unlikely that the low numbers of transcripts measured represent ubiquitously distributed, rare nuclear RNAs that may not ever be processed in the embryo (33), which are present at any given time at levels of 0.05–1 molecule per cell (22).
We conclude that none of the embryo-wide specification functions, and in the limit none but the finishing touches of embryonic morphogenesis, could possibly require most of the genes of the Hox cluster. Yet, the pluteus-stage larva, as shown in Fig. 2A, is in fact a bilaterally organized, free-living metazoan, equipped with differentiated mesodermal, neuroectodermal, and endodermal cell types. It is a very small metazoan, in which lineage relationships organized with respect to the cytoarchitecture of the egg, and direct short-range signaling interactions, suffice for its development. It is not true, therefore, that the Hox cluster is a definitive and essential component of the genetic regulatory apparatus that underlies development of every free-living metazoan organism of bilaterian grade, although it is likely to be essential for the development of the adult body plans of all macroscopic bilaterian organisms.
Indirect Development, Hox Genes, and Metazoan Evolution.
We have discussed elsewhere evidence that has led us and our colleagues to conclude that indirect development is primitive for bilaterians (1, 5). Basically there are two arguments: first, the very widespread phylogenetic distribution of indirect development, both in deuterostomes and in lophotrochozoan protostomes; and second, the remarkably similar morphological organization of larvae that nonetheless give rise to very different adult body plans. For example, in deuterostomes, echinoderms and enteropneust hemichordate larvae display an uncanny similarity in structure, particularly in the tripartite disposition of their coelomic mesodermal set-aside cells, which ultimately generate major portions of the entirely different adult body plans of these animals. Similar examples abound in lophotrochozoans (1). Strikingly, molecular phylogeny confirms the relations implied by the homologies among deuterostome and protostome larval structures, respectively, despite the very different phyletic adult body plans emerging within each clade. It does not seem likely that these larval forms are the convergent results of independent derivations from more primitive direct developing forms, given the details of similarity in larval structure observed both among deuterostome dipleurula larvae and among lophotrochozoan trochophore larvae. We conclude that the correct evolutionary polarity is the reverse: indirect development is basal for these clades, and their diverse adult body plans evolved from a much less diverse, prior micrometazoan fauna, but of bilaterian grade of organization. The present work adds to the discussion of metazoan origins experimental evidence that the genetic regulatory apparatus needed to build such micrometazoan organisms is indeed qualitatively simpler than that needed for the development of any known adult bilaterian body plan. This fact lends indirect support to the argument (5) that for a long period of metazoan evolution there could have existed ancestral bilaterian forms, the development of which did not require the complex regulatory apparatus used in modern organisms for spatial specification of large populations of growing cells. In this light, maximal indirect development can be seen, in many different bilaterian clades, as a developmental succession of distinct regulatory functions that represent two major evolutionary innovations. These might have appeared far apart in time. Regulatory utilization of all of the genes of the Hox gene complex appears now as an integral aspect of the later set of regulatory inventions, that leading to the adult body plans of large animals. To extend this argument beyond deuterostomes, it is obviously now of immediate interest to measure Hox gene utilization in an indirectly developing lophotrochozoan embryo/larva. These animals have a homologous set of Hox genes (34), and it is important to determine whether they display the same pattern of developmental Hox gene utilization, despite the deep evolutionary divergence that separates them from the deuterostomes.
Acknowledgments
We are grateful to Prof. André Adoutte (Université de Paris-Sud) and to Prof. Ellen Rothenberg and Dr. Kevin Peterson (this institute) for reviews and discussions of this manuscript. This work was supported by the Stowers Institute for Medical Research and by National Institutes of Health Grant HD05753 (to E.H.D.) and from the National Science Foundation Grant IBN9604454 (to R.A.C.).
ABBREVIATION
- WMISH
whole-mount in situ hybridization
References
- 1. Peterson K J, Cameron R A, Davidson E H. BioEssays. 1997;19:623–631. doi: 10.1002/bies.950190713. [DOI] [PubMed] [Google Scholar]
- 2.Salser S J, Kenyon C. Trends Genet. 1994;10:159–164. doi: 10.1016/0168-9525(94)90092-2. [DOI] [PubMed] [Google Scholar]
- 3.Kourakis M J, Master V A, Lokhorst D K, Nardelli-Haefliger D, Weeden C J, Martindale M Q, Shankland M. Dev Biol. 1997;190:284–300. doi: 10.1006/dbio.1997.8689. [DOI] [PubMed] [Google Scholar]
- 4.Wong V Y, Aisemberg G O, Macagno E R. J Neurosci. 1995;15:5551–5559. doi: 10.1523/JNEUROSCI.15-08-05551.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davidson E H, Peterson K, Cameron R A. Science. 1995;270:1319–1325. doi: 10.1126/science.270.5240.1319. [DOI] [PubMed] [Google Scholar]
- 6.Davidson E H. Development (Cambridge, UK) 1990;108:365–389. doi: 10.1242/dev.108.3.365. [DOI] [PubMed] [Google Scholar]
- 7.Davidson E H. Development (Cambridge, UK) 1991;113:1–26. doi: 10.1242/dev.113.1.1. [DOI] [PubMed] [Google Scholar]
- 8.Halanych K M, Bacheller J D, Aquilnaldo A M A, Liva S M, Hillis D M, Lake J A. Science. 1995;267:1641–1643. doi: 10.1126/science.7886451. [DOI] [PubMed] [Google Scholar]
- 9.Davidson E H. BioEssays. 1994;16:603–615. doi: 10.1002/bies.950160903. [DOI] [PubMed] [Google Scholar]
- 10.Williams J A, Paddock S W, Carroll S B. Development (Cambridge, UK) 1993;117:571–584. doi: 10.1242/dev.117.2.571. [DOI] [PubMed] [Google Scholar]
- 11.Johnson R L, Tabin C J. Cell. 1997;90:979–990. doi: 10.1016/s0092-8674(00)80364-5. [DOI] [PubMed] [Google Scholar]
- 12.Bally-Cuif L, Boncinelli E. BioEssays. 1996;19:127–135. doi: 10.1002/bies.950190207. [DOI] [PubMed] [Google Scholar]
- 13.Harland R, Gerhart J. Annu Rev Cell Dev. 1997;13:611–677. doi: 10.1146/annurev.cellbio.13.1.611. [DOI] [PubMed] [Google Scholar]
- 14.McGinnis W, Krumlauf R. Cell. 1992;68:283–302. doi: 10.1016/0092-8674(92)90471-n. [DOI] [PubMed] [Google Scholar]
- 15.Carroll S B. Nature (London) 1995;376:479–485. doi: 10.1038/376479a0. [DOI] [PubMed] [Google Scholar]
- 16.Carroll S B. Genes Dev. 1998;12:1474–1482. doi: 10.1101/gad.12.10.1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Akam M. Int J Dev Biol. 1998;42:445–451. [PubMed] [Google Scholar]
- 18.Wang B B, Mueller-Immergluck M M, Austin J, Robinson N T, Chisholm A, Kenyon C. Cell. 1993;74:29–42. doi: 10.1016/0092-8674(93)90292-x. [DOI] [PubMed] [Google Scholar]
- 19.Nardelli-Haefliger D, Bruce A E E, Shankland M. Development (Cambridge, UK) 1994;120:1839–1849. doi: 10.1242/dev.120.7.1839. [DOI] [PubMed] [Google Scholar]
- 20.Lee J J, Costlow N A. Methods Enzymol. 1987;152:633–648. doi: 10.1016/0076-6879(87)52070-5. [DOI] [PubMed] [Google Scholar]
- 21.Holland L Z, Holland P W H, Holland N D. In: Molecular Zoology. Ferraris J D, Palumbi S R, editors. New York: Wiley–Liss; 1996. pp. 267–281. [Google Scholar]
- 22.Davidson E H. Gene Activity in Early Development. 3rd Ed. Orlando, FL: Academic; 1986. [Google Scholar]
- 23.Angerer L M, Dolecki G J, Gagnon M, Lum R, Wang G, Yang Q, Humphreys T, Angerer R C. Genes Dev. 1989;3:370–383. doi: 10.1101/gad.3.3.370. [DOI] [PubMed] [Google Scholar]
- 24.Dobias S L, Zhao A Z, Tan H, Bell J R, Maxson R. Dev Dyn. 1996;207:450–460. doi: 10.1002/(SICI)1097-0177(199612)207:4<450::AID-AJA9>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 25.Wang D G-W, Britten R J, Davidson E H. Mol Marine Biol Biotechnol. 1995;4:148–153. [PubMed] [Google Scholar]
- 26.Cameron R A, Britten R J, Davidson E H. Mol Reprod Dev. 1989;1:149–155. doi: 10.1002/mrd.1080010302. [DOI] [PubMed] [Google Scholar]
- 27.Lee J J, Shott R J, Rose S J, Thomas T L, Britten R J, Davidson E H. J Mol Biol. 1984;172:149–176. doi: 10.1016/s0022-2836(84)80035-2. [DOI] [PubMed] [Google Scholar]
- 28.Davidson E H, Cameron R A, Ransick A. Development (Cambridge, UK) 1998;125:3269–3290. doi: 10.1242/dev.125.17.3269. [DOI] [PubMed] [Google Scholar]
- 29.Hyman L H. The Invertebrates: Echinodermata. IV. New York: McGraw-Hill; 1955. [Google Scholar]
- 30.Smith L C, Davidson E H. Ann NY Acad Sci. 1994;712:213–226. doi: 10.1111/j.1749-6632.1994.tb33575.x. [DOI] [PubMed] [Google Scholar]
- 31.Cameron R A, Hough-Evans B R, Britten R J, Davidson E H. Genes Dev. 1987;1:75–85. doi: 10.1101/gad.1.1.75. [DOI] [PubMed] [Google Scholar]
- 32.Zhao A Z, Vansant G, Bell J, Humphreys T, Maxson R. Mech Dev. 1991;34:21–28. doi: 10.1016/0925-4773(91)90088-n. [DOI] [PubMed] [Google Scholar]
- 33.Wold B J, Klein W H, Hough-Evans B R, Britten R J, Davidson E H. Cell. 1978;14:941–950. doi: 10.1016/0092-8674(78)90348-3. [DOI] [PubMed] [Google Scholar]
- 34.Adoutte, A. (1998) Trends Genet., in press. [DOI] [PubMed]