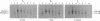Abstract
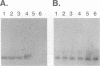
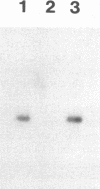
Competitive quantitative PCR and reverse transcriptase-PCR were used to quantitate DNA and RNA from an attenuated ribonucleotide reductase-deleted herpes simplex virus type 1 (HSV-1) mutant in the rat trigeminal ganglion after peripheral inoculation following corneal scarification. Amplification of ganglionic DNA with oligonucleotide primers specific for the HSV-1 glycoprotein B (gB) gene and for the latency-associated transcript (LAT) gene indicated that there were approximately 2 x 10(5) genome equivalents per ganglion at 2 days, 7 days, and 8 weeks after inoculation. Amplification of ganglionic RNA with primers specific for HSV-1 LAT indicated that the amount of LAT RNA was also stable over 8 weeks, with 10(7) LAT molecules per ganglion at 2 days and at 7 days postinoculation and 1.4 x 10(7) LAT molecules per ganglion at 8 weeks. In situ hybridization with a digoxigenin-labeled riboprobe specific for LAT detected an average of one to two LAT-positive cells in each positive 6-microns section of trigeminal ganglion. In situ PCR detection of HSV-1 genomes in similar sections, using digoxigenin-labeled nucleotides with primers specific for HSV-1 gB, identified as many as 120 genome-positive cells per section. These results indicate that there are approximately 50 LAT molecules per latent HSV-1 genome in the trigeminal ganglion, compared with 15 LAT molecules per latent HSV-1 genome in the central nervous system (R. Ramakrishnan, D. J. Fink, G. Jiang, P. Desai, J. C. Glorioso, and M. Levine, J. Virol. 68:1864-1873, 1994), but that cells with detectable LATs by in situ hybridization represent only a small proportion of those ganglionic neurons containing HSV-1 genomes. The presence of latent HSV-1 genomes in a large number of neurons suggests that HSV-1 may be more efficient in establishing the latent state than would be anticipated from previous reports.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arvidson B. Retrograde axonal transport of horseradish peroxidase from cornea to trigeminal ganglion. Acta Neuropathol. 1977 Apr 29;38(1):49–52. doi: 10.1007/BF00691276. [DOI] [PubMed] [Google Scholar]
- Becker-André M., Hahlbrock K. Absolute mRNA quantification using the polymerase chain reaction (PCR). A novel approach by a PCR aided transcript titration assay (PATTY). Nucleic Acids Res. 1989 Nov 25;17(22):9437–9446. doi: 10.1093/nar/17.22.9437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block T. M., Deshmane S., Masonis J., Maggioncalda J., Valyi-Nagi T., Fraser N. W. An HSV LAT null mutant reactivates slowly from latent infection and makes small plaques on CV-1 monolayers. Virology. 1993 Feb;192(2):618–630. doi: 10.1006/viro.1993.1078. [DOI] [PubMed] [Google Scholar]
- Cai W., Astor T. L., Liptak L. M., Cho C., Coen D. M., Schaffer P. A. The herpes simplex virus type 1 regulatory protein ICP0 enhances virus replication during acute infection and reactivation from latency. J Virol. 1993 Dec;67(12):7501–7512. doi: 10.1128/jvi.67.12.7501-7512.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron J. M., McDougall I., Marsden H. S., Preston V. G., Ryan D. M., Subak-Sharpe J. H. Ribonucleotide reductase encoded by herpes simplex virus is a determinant of the pathogenicity of the virus in mice and a valid antiviral target. J Gen Virol. 1988 Oct;69(Pt 10):2607–2612. doi: 10.1099/0022-1317-69-10-2607. [DOI] [PubMed] [Google Scholar]
- Coen D. M., Kosz-Vnenchak M., Jacobson J. G., Leib D. A., Bogard C. L., Schaffer P. A., Tyler K. L., Knipe D. M. Thymidine kinase-negative herpes simplex virus mutants establish latency in mouse trigeminal ganglia but do not reactivate. Proc Natl Acad Sci U S A. 1989 Jun;86(12):4736–4740. doi: 10.1073/pnas.86.12.4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deatly A. M., Spivack J. G., Lavi E., O'Boyle D. R., 2nd, Fraser N. W. Latent herpes simplex virus type 1 transcripts in peripheral and central nervous system tissues of mice map to similar regions of the viral genome. J Virol. 1988 Mar;62(3):749–756. doi: 10.1128/jvi.62.3.749-756.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai P., Ramakrishnan R., Lin Z. W., Osak B., Glorioso J. C., Levine M. The RR1 gene of herpes simplex virus type 1 is uniquely trans activated by ICP0 during infection. J Virol. 1993 Oct;67(10):6125–6135. doi: 10.1128/jvi.67.10.6125-6135.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshmane S. L., Fraser N. W. During latency, herpes simplex virus type 1 DNA is associated with nucleosomes in a chromatin structure. J Virol. 1989 Feb;63(2):943–947. doi: 10.1128/jvi.63.2.943-947.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshmane S. L., Nicosia M., Valyi-Nagy T., Feldman L. T., Dillner A., Fraser N. W. An HSV-1 mutant lacking the LAT TATA element reactivates normally in explant cocultivation. Virology. 1993 Oct;196(2):868–872. doi: 10.1006/viro.1993.1548. [DOI] [PubMed] [Google Scholar]
- Devi-Rao G. B., Goodart S. A., Hecht L. M., Rochford R., Rice M. K., Wagner E. K. Relationship between polyadenylated and nonpolyadenylated herpes simplex virus type 1 latency-associated transcripts. J Virol. 1991 May;65(5):2179–2190. doi: 10.1128/jvi.65.5.2179-2190.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efstathiou S., Minson A. C., Field H. J., Anderson J. R., Wildy P. Detection of herpes simplex virus-specific DNA sequences in latently infected mice and in humans. J Virol. 1986 Feb;57(2):446–455. doi: 10.1128/jvi.57.2.446-455.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink D. J., Sternberg L. R., Weber P. C., Mata M., Goins W. F., Glorioso J. C. In vivo expression of beta-galactosidase in hippocampal neurons by HSV-mediated gene transfer. Hum Gene Ther. 1992 Feb;3(1):11–19. doi: 10.1089/hum.1992.3.1-11. [DOI] [PubMed] [Google Scholar]
- Fraser N. W., Block T. M., Spivack J. G. The latency-associated transcripts of herpes simplex virus: RNA in search of function. Virology. 1992 Nov;191(1):1–8. doi: 10.1016/0042-6822(92)90160-q. [DOI] [PubMed] [Google Scholar]
- Gordon Y. J., Johnson B., Romanowski E., Araullo-Cruz T. RNA complementary to herpes simplex virus type 1 ICP0 gene demonstrated in neurons of human trigeminal ganglia. J Virol. 1988 May;62(5):1832–1835. doi: 10.1128/jvi.62.5.1832-1835.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gressens P., Martin J. R. In situ polymerase chain reaction: localization of HSV-2 DNA sequences in infections of the nervous system. J Virol Methods. 1994 Jan;46(1):61–83. doi: 10.1016/0166-0934(94)90017-5. [DOI] [PubMed] [Google Scholar]
- Ho D. Y., Mocarski E. S. Herpes simplex virus latent RNA (LAT) is not required for latent infection in the mouse. Proc Natl Acad Sci U S A. 1989 Oct;86(19):7596–7600. doi: 10.1073/pnas.86.19.7596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson J. G., Leib D. A., Goldstein D. J., Bogard C. L., Schaffer P. A., Weller S. K., Coen D. M. A herpes simplex virus ribonucleotide reductase deletion mutant is defective for productive acute and reactivatable latent infections of mice and for replication in mouse cells. Virology. 1989 Nov;173(1):276–283. doi: 10.1016/0042-6822(89)90244-4. [DOI] [PubMed] [Google Scholar]
- Katz J. P., Bodin E. T., Coen D. M. Quantitative polymerase chain reaction analysis of herpes simplex virus DNA in ganglia of mice infected with replication-incompetent mutants. J Virol. 1990 Sep;64(9):4288–4295. doi: 10.1128/jvi.64.9.4288-4295.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leib D. A., Bogard C. L., Kosz-Vnenchak M., Hicks K. A., Coen D. M., Knipe D. M., Schaffer P. A. A deletion mutant of the latency-associated transcript of herpes simplex virus type 1 reactivates from the latent state with reduced frequency. J Virol. 1989 Jul;63(7):2893–2900. doi: 10.1128/jvi.63.7.2893-2900.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynas C., Laycock K. A., Cook S. D., Hill T. J., Blyth W. A., Maitland N. J. Detection of herpes simplex virus type 1 gene expression in latently and productively infected mouse ganglia using the polymerase chain reaction. J Gen Virol. 1989 Sep;70(Pt 9):2345–2355. doi: 10.1099/0022-1317-70-9-2345. [DOI] [PubMed] [Google Scholar]
- Marfurt C. F., Kingsley R. E., Echtenkamp S. E. Sensory and sympathetic innervation of the mammalian cornea. A retrograde tracing study. Invest Ophthalmol Vis Sci. 1989 Mar;30(3):461–472. [PubMed] [Google Scholar]
- Margolis T. P., Dawson C. R., LaVail J. H. Herpes simplex viral infection of the mouse trigeminal ganglion. Immunohistochemical analysis of cell populations. Invest Ophthalmol Vis Sci. 1992 Feb;33(2):259–267. [PubMed] [Google Scholar]
- Mellerick D. M., Fraser N. W. Physical state of the latent herpes simplex virus genome in a mouse model system: evidence suggesting an episomal state. Virology. 1987 Jun;158(2):265–275. doi: 10.1016/0042-6822(87)90198-x. [DOI] [PubMed] [Google Scholar]
- Mitchell W. J., Lirette R. P., Fraser N. W. Mapping of low abundance latency-associated RNA in the trigeminal ganglia of mice latently infected with herpes simplex virus type 1. J Gen Virol. 1990 Jan;71(Pt 1):125–132. doi: 10.1099/0022-1317-71-1-125. [DOI] [PubMed] [Google Scholar]
- Puga A., Notkins A. L. Continued expression of a poly(A)+ transcript of herpes simplex virus type 1 in trigeminal ganglia of latently infected mice. J Virol. 1987 May;61(5):1700–1703. doi: 10.1128/jvi.61.5.1700-1703.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan R., Fink D. J., Jiang G., Desai P., Glorioso J. C., Levine M. Competitive quantitative PCR analysis of herpes simplex virus type 1 DNA and latency-associated transcript RNA in latently infected cells of the rat brain. J Virol. 1994 Mar;68(3):1864–1873. doi: 10.1128/jvi.68.3.1864-1873.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock D. L., Fraser N. W. Detection of HSV-1 genome in central nervous system of latently infected mice. Nature. 1983 Apr 7;302(5908):523–525. doi: 10.1038/302523a0. [DOI] [PubMed] [Google Scholar]
- Rock D. L., Nesburn A. B., Ghiasi H., Ong J., Lewis T. L., Lokensgard J. R., Wechsler S. L. Detection of latency-related viral RNAs in trigeminal ganglia of rabbits latently infected with herpes simplex virus type 1. J Virol. 1987 Dec;61(12):3820–3826. doi: 10.1128/jvi.61.12.3820-3826.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roizman B., Sears A. E. An inquiry into the mechanisms of herpes simplex virus latency. Annu Rev Microbiol. 1987;41:543–571. doi: 10.1146/annurev.mi.41.100187.002551. [DOI] [PubMed] [Google Scholar]
- Rødahl E., Stevens J. G. Differential accumulation of herpes simplex virus type 1 latency-associated transcripts in sensory and autonomic ganglia. Virology. 1992 Jul;189(1):385–388. doi: 10.1016/0042-6822(92)90721-z. [DOI] [PubMed] [Google Scholar]
- Sawtell N. M., Thompson R. L. Herpes simplex virus type 1 latency-associated transcription unit promotes anatomical site-dependent establishment and reactivation from latency. J Virol. 1992 Apr;66(4):2157–2169. doi: 10.1128/jvi.66.4.2157-2169.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedarati F., Izumi K. M., Wagner E. K., Stevens J. G. Herpes simplex virus type 1 latency-associated transcription plays no role in establishment or maintenance of a latent infection in murine sensory neurons. J Virol. 1989 Oct;63(10):4455–4458. doi: 10.1128/jvi.63.10.4455-4458.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spivack J. G., Fraser N. W. Detection of herpes simplex virus type 1 transcripts during latent infection in mice. J Virol. 1987 Dec;61(12):3841–3847. doi: 10.1128/jvi.61.12.3841-3847.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staskus K. A., Couch L., Bitterman P., Retzel E. F., Zupancic M., List J., Haase A. T. In situ amplification of visna virus DNA in tissue sections reveals a reservoir of latently infected cells. Microb Pathog. 1991 Jul;11(1):67–76. doi: 10.1016/0882-4010(91)90095-r. [DOI] [PubMed] [Google Scholar]
- Steiner I., Spivack J. G., Lirette R. P., Brown S. M., MacLean A. R., Subak-Sharpe J. H., Fraser N. W. Herpes simplex virus type 1 latency-associated transcripts are evidently not essential for latent infection. EMBO J. 1989 Feb;8(2):505–511. doi: 10.1002/j.1460-2075.1989.tb03404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens J. G., Cook M. L. Latent herpes simplex virus in spinal ganglia of mice. Science. 1971 Aug 27;173(3999):843–845. doi: 10.1126/science.173.3999.843. [DOI] [PubMed] [Google Scholar]
- Stevens J. G. Human herpesviruses: a consideration of the latent state. Microbiol Rev. 1989 Sep;53(3):318–332. doi: 10.1128/mr.53.3.318-332.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens J. G., Wagner E. K., Devi-Rao G. B., Cook M. L., Feldman L. T. RNA complementary to a herpesvirus alpha gene mRNA is prominent in latently infected neurons. Science. 1987 Feb 27;235(4792):1056–1059. doi: 10.1126/science.2434993. [DOI] [PubMed] [Google Scholar]
- Wagner E. K., Devi-Rao G., Feldman L. T., Dobson A. T., Zhang Y. F., Flanagan W. M., Stevens J. G. Physical characterization of the herpes simplex virus latency-associated transcript in neurons. J Virol. 1988 Apr;62(4):1194–1202. doi: 10.1128/jvi.62.4.1194-1202.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner E. K., Flanagan W. M., Devi-Rao G., Zhang Y. F., Hill J. M., Anderson K. P., Stevens J. G. The herpes simplex virus latency-associated transcript is spliced during the latent phase of infection. J Virol. 1988 Dec;62(12):4577–4585. doi: 10.1128/jvi.62.12.4577-4585.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wechsler S. L., Nesburn A. B., Watson R., Slanina S. M., Ghiasi H. Fine mapping of the latency-related gene of herpes simplex virus type 1: alternative splicing produces distinct latency-related RNAs containing open reading frames. J Virol. 1988 Nov;62(11):4051–4058. doi: 10.1128/jvi.62.11.4051-4058.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]