Abstract
Fanconi anemia (FA) is an autosomal recessive cancer susceptibility syndrome with at least eight complementation groups (A–H). Two FA genes, corresponding to complementation groups A and C, have been cloned, but the function of the FAA and FAC proteins remains unknown. We have recently shown that the FAA and FAC proteins bind and form a nuclear complex. In the current study, we analyzed the FAA and FAC proteins in normal lymphoblasts and lymphoblasts from multiple FA complementation groups. In contrast to normal controls, FA cells derived from groups A, B, C, E, F, G, and H were defective in the formation of the FAA/FAC protein complex, the phosphorylation of the FAA protein, and the accumulation of the FAA/FAC protein complex in the nucleus. These biochemical events seem to define a signaling pathway required for the maintenance of genomic stability and normal hematopoiesis. Our results support the idea that multiple gene products cooperate in the FA Pathway.
Keywords: mitomycin C/leukemia/cancer susceptibility
Fanconi anemia (FA) is an autosomal recessive disease characterized by genomic instability, cancer susceptibility, and progressive bone marrow failure (refs. 1–3; for ref. 3 see section, “Fanconi anemia”). Somatic cell fusion studies have defined at least eight genetic complementation groups (FA-A through FA-H; refs. 4–6), displaying similar phenotypes, suggesting a functional relationship of the FA genes. The genes corresponding to FA-A and FA-C have been cloned (7–9), and mutations in FAA and FAC account for 80% of patients with FA (6, 10). The FAA and FAC proteins have no sequence similarity to other proteins in GenBank, and their biochemical functions remain unknown. We have recently determined that FAA and FAC bind and form a protein complex in the nucleus (11). The FA proteins may therefore cooperate in some nuclear function, such as DNA repair, DNA replication, or RNA splicing.
Cells derived from patients with FA display multiple phenotypic abnormalities. FA cells are hypersensitive to bifunctional alkylating agents, such as diepoxybutane and mitomycin C (MMC), suggesting a defect in DNA repair. FA cells also exhibit abnormal cell-cycle progression (12–14) and reduced cell survival (15–19). Many of these abnormalities are also evident in primary cells derived from mice homozygous for a disrupted fac gene (16, 20). Currently, how the absence of the FAA protein, the FAC protein, or the FAA/FAC protein complex leads to these cellular abnormalities is unknown.
Little is known regarding the nature of the binding interaction between the FAA and FAC proteins. The binding may be a direct protein–protein interaction or may be an indirect interaction, mediated by other adaptor proteins. Regulated posttranslational modifications of the FAA or FAC protein, such as phosphorylation, may also be required for interaction of the FA proteins. It is not yet known whether the FAA/FAC binding interaction is constitutive or inducible under various cellular conditions or stresses.
Interaction between the FAA and FAC proteins has functional importance. A patient-derived missense mutation in the FAC protein, FACL554P, prevents the formation of the FAA/FAC complex (11), suggesting that protein binding may be required for the normal (non-FA) cellular phenotype. Whether the interaction of FAA and FAC is required for nuclear translocation of the protein complex also is currently unclear. The expression and binding of FAA and FAC in the various FA complementation groups has not yet been examined.
To explore the functional importance of the FAA/FAC protein interaction further, we analyzed FAA/FAC binding, FAA phosphorylation, and FAA/FAC nuclear accumulation in lymphoblast lines derived from normal adult controls or patients with FA. Our results show that the FAA protein is phosphorylated in normal cells and FA-D cells but not in FA cells derived from groups A, B, C, E, F, G, and H. Moreover, the phosphorylation of FAA correlated with both the binding of FAA to FAC and the nuclear accumulation of FAA and FAC. Our results suggest that FAA phosphorylation is an important regulatory event controlling the FA signaling pathway.
MATERIALS AND METHODS
Characterization of Cell Lines.
FA lymphoblast lines and PD7 cells (normal control) were cultured as described (21–23). All FA cell lines used in this study were sensitive to MMC as described (6, 7, 11). FA lymphoblast lines were also analyzed by transduction with retroviral vectors containing FAA or FAC cDNAs (24). As expected, FAA and FAC cDNAs failed to complement the MMC sensitivity of FA cells derived from groups B, D, E, F, G, and H.
Production of pMMP-FAA and pMMP-FAC Retroviral Supernatants and Infection of Lymphoblast Lines.
The FAA cDNA or FAC cDNA was subcloned into the retroviral vector, pMMP (25), as described (11, 24). For production of pMMP-FAA or pMMP-FAC retrovirus, 293GPG helper cells (25) were grown to 90% confluence in 10-cm dishes and electroporated by lipofection for 8 h at 37°C with 10 μg of plasmid DNA in 6 ml of Opti-MEM (GIBCO) containing 8.7 ml/ml Lipofectamine (GIBCO). Fresh medium (10 ml of DMEM/15% fetal calf serum) was added and replaced every 24 h. Virus-containing medium supernatants were harvested 96, 120, and 144 h after lipofection and clarified by filtration (0.45 μm). Viral supernatants (5 ml) were mixed with an equal volume of RPMI medium 1640 with 15% fetal calf serum containing 8 μg/ml hexadimethrine bromide (Sigma) and added to six-well plates containing 5 × 104 lymphoblasts per well. After incubation for 4–6 h at 37°C in 5% CO2, the medium was replaced with RPMI medium 1640 with 15% fetal calf serum. Retrovirally transduced lymphoblasts were analyzed by the MMC assay as described (21).
Western Blotting and Immunoprecipitation.
Western blotting and immunoprecipitation of FAA and FAC were performed with affinity-purified polyclonal rabbit antisera as described (11). The anti-FAA(N) and anti-FAA(C) antisera were raised against the N terminus or the C terminus of FAA, respectively.
In Vivo Labeling with [32P]Orthophosphate.
Lymphoblasts (≈1 × 107 cells per sample) were preincubated in phosphate-free medium for 30 min, then incubated in medium containing 1 mCi/ml [32P]orthophosphate for 2 h. Cells were washed twice with ice-cold PBS, then lysed in 1 ml of lysis buffer (50 mM Tris⋅HCl, pH 7.5/100 mM NaCl/30 mM sodium pyrophosphate/1 mM sodium orthovanadate/5 mM EDTA/1% Triton X-100/1 mM phenylmethylsulfonyl fluoride/10 μg/ml leupeptin/2 μg/ml aprotinin). Nuclei and debris were removed by centrifugation for 20 min in a microcentrifuge tube. Immunoprecipitation with a 1:1 mixture of anti-FAA(N) and anti-FAA(C) antisera was performed in cell lysates containing 0.2% SDS and 5 mg/ml sodium cholate. The protein A-bound immunoprecipitates were washed extensively with lysis buffer containing 0.1% SDS and boiled in 50 μl of SDS-loading buffer. Denatured proteins were separated on SDS/polyacrylamide gels.
Lymphoblast Fractionation.
For fractionation of lymphoblasts, cells were washed in PBS, resuspended in buffer (10 mM Tris, pH 7.4/3 mM CaCl2/2 mM MgCl2/1% Nonidet P-40), and lysed in a Dounce homogenizer with 30 strokes. Nuclei were pelleted by centrifugation at 1,500 rpm in a model RC3B centrifuge (Sorvall), and the supernatant (cytosolic fraction) was clarified by centrifugation at 15,000 rpm. Nuclei were washed three times with PBS and lysed in buffer (50 mM Tris⋅HCl, pH 7.4/150 mM NaCl/1% Triton X-100). Nuclear lysates were clarified by centrifugation at 15,000 rpm in a microcentrifuge.
RESULTS
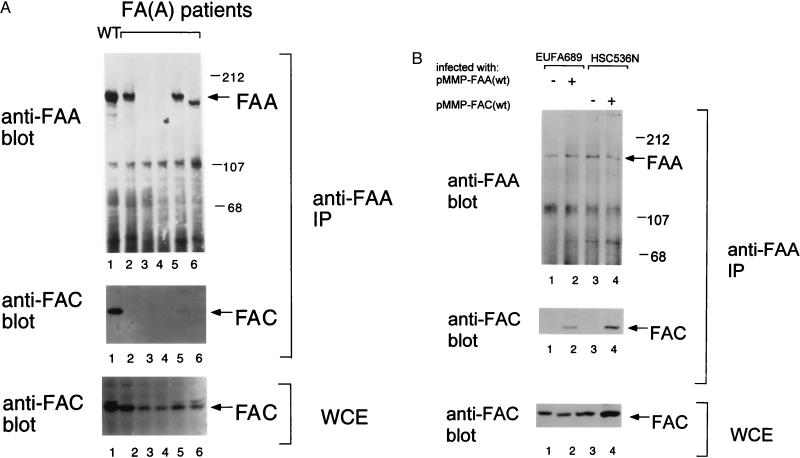
Initially, we examined the interaction of FAA and FAC proteins in normal and FA lymphoblast lines (Fig. 1). For the normal control cell line PD7, wt FAA protein coimmunoprecipitated with wt FAC protein (Fig. 1A, lane 1) as described (11). Interestingly, in cells expressing a mutant FAA protein, FAA(delF1263) (26, 27), FAA did not coimmunoprecipitate or only weakly immunoprecipitated with FAC (Fig. 1A, lanes 2 and 5). Also, for the FA-A lines, HSC72 (Fig. 1A, lane 3) and PD9 (Fig. 1A, lane 4), no FAA protein was observed, and the FAC protein did not immunoprecipitate with the anti-FAA antibody. Another FA-A lymphoblast line, PD475, expressed a truncated FAA protein that failed to bind FAC (Fig. 1A, lane 6). FAC was expressed at comparable levels in all cell lines examined (whole-cell lysates, FAC immunoblotting). The absence of FAA/FAC binding was therefore common to all FA-A cell lines examined, thereby confirming the functional importance of the protein complex.
Figure 1.
Mutant FAA proteins, expressed in patient-derived FA-A cell lines, fail to bind to wild-type (wt) FAC protein. (A) Epstein–Barr-virus-transformed lymphoblast lines, derived from a normal-adult control or from five known patients with FA-A, were analyzed for the expression of FAA and FAC protein. Protein from the indicated cell lines was immunoprecipitated (IP) with an anti-FAA (C-terminal) antiserum. Protein was electrophoresed, transferred to nitrocellulose, and immunoblotted with either anti-FAA (N-terminal) or anti-FAC antiserum. Alternatively, whole-cell extracts (WCE) were directly analyzed by anti-FAC immunoblot (Bottom). Cell lines analyzed included the normal control PD7 (lane 1) and the MMC-sensitive FA-A cell lines, EUFA689 (lane 2), HSC72 (lane 3), PD9 (lane 4), EUFA006 (lane 5), or PD475 (lane 6; ref. 11). The EUFA689 cell line is homozygous for the 3788–3790del FAA allele, whereas the EUFA006 cell line is a compound heterozygote containing the 3788–3790del FAA allele and the 1391del467FAA allele (27). The HSC72 cell line has a homozygous deletion of the FAA gene (H.J., unpublished results). The genotypes of the other FA-A cell lines (PD9 and PD475) have not been determined. (B) Binding of FAA and FAC protein in functionally complemented FA-A and FA-C cell lines. The MMC sensitivity of EUFA689 (FA-A) cells and HSC536 (FA-C) cells was corrected by retroviral transduction with the FAA cDNA and FAC cDNA, respectively (data not shown). Cellular proteins from the indicated cell lines were immunoprecipitated with an anti-FAA (C-terminal) antiserum. Alternatively, whole-cell extracts were directly analyzed by anti-FAC immunoblots (Bottom). Parental HSC536 cells express the mutant FAC protein (FACL554P), which has been described (7, 37).
We next analyzed functionally complemented FA cell lines for the physical interaction of FAA and FAC protein (Fig. 1B). The MMC sensitivity of EUFA689 (FA-A) cells and HSC536 (FA-C) cells was corrected by retroviral transduction with the FAA cDNA or FAC cDNA, respectively (data not shown). In EUFA689/pMMP-FAA (wt) cells, the wt FAA protein coimmunoprecipitated with endogenous FAC protein (Fig. 1B, lane 2), indicating further that FAC binding correlates with the functional activity of FAA. Similarly, in HSC536/pMMP-FAC (wt) cells, the wt FAC protein coimmunoprecipitated with endogenous FAA protein (Fig. 1B, lane 4).
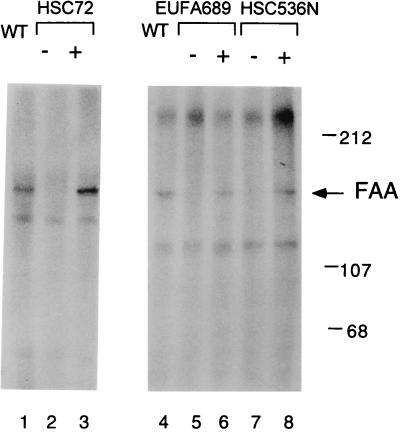
To determine whether FA proteins are phosphorylated, we labeled the various lymphoblast lines with [32P]orthophosphate and immunoprecipitated the FAA protein (Fig. 2). A 163-kDa phosphoprotein (phosphorylated FAA) immunoprecipitated with the anti-FAA antiserum from extracts of wt PD7 cells (Fig. 2, lanes 1 and 4) and functionally complemented cells (Fig. 2, lanes 3, 6, and 8). By phosphoamino acid analysis, FAA was phosphorylated primarily on serine residues (data not shown). The mutant FAA protein, FAAdelF1263, was not phosphorylated in EUFA689 cells (Fig. 2, lane 5). Also, the wt FAA protein was not phosphorylated in cells expressing mutant FAC protein (HSC536 cells; Fig. 2, lane 7). No phosphorylated FAA protein was detected in HSC72 cells (Fig. 2, lane 2), consistent with the absence of FAA in these cells determined by immunoblot analysis. Phosphorylated FAA was detected in HSC72/pMMP-FAA cells (Fig. 2, lane 3). Phosphorylated FAC protein was not detected in any cell lines, suggesting that FAC is not a phosphoprotein (data not shown). Taken together, these data show that FAA is a phosphoprotein whose phosphorylation correlates with the functional complementation (MMC resistance) of each cell line.
Figure 2.
Phosphorylation of the FAA protein correlates with the function of the FA pathway. The indicated human lymphoblast lines were labeled in vivo by incubation with [32P]orthophosphate. Cell extracts were immunoprecipitated with an anti-FAA (C-terminal) antiserum and separated by denaturing PAGE. Gels were dried, and radiolabeled proteins were visualized by autoradiography for 12 to 24 h at −70°C. Cell lines examined were PD7 (lanes 1, 4), HSC72 (lane 2), HSC72 cells corrected with FAA (lane 3), EUFA689 (lane 5), EUFA689 corrected with FAA (lane 6), HSC536 (lane 7), and HSC536 corrected with FAC (lane 8).
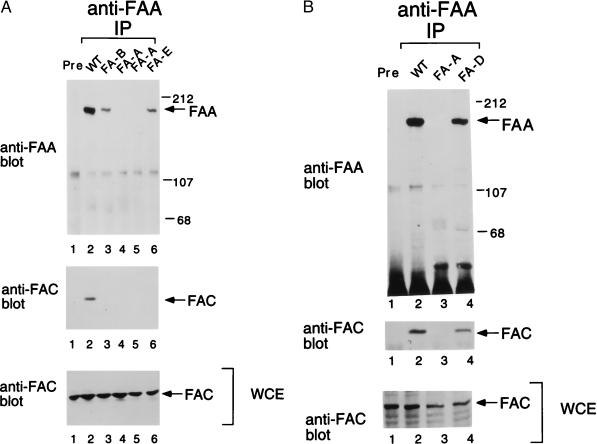
We next analyzed FAA/FAC binding in lymphoblast lines derived from patients with FA of other complementation groups (Fig. 3). Like FA-A and FA-C lymphoblasts, FA-B, FA-D, and FA-E lymphoblasts were sensitive to MMC. FA-B, FA-D, and FA-E lymphoblast lines expressed normal levels of FAA and FAC protein (Fig. 3 A, lanes 3 and 6, and B, lane 4) and were not complemented by stable transfection with the FAA or FAC cDNAs (data not shown). The FAA and FAC proteins coimmunoprecipitated from normal cells (Fig. 3A, lane 2) and FA-D cells (Fig. 3B, lane 4), but not from FA-B and FA-E cells (Fig. 3A, lanes 3 and 6, respectively). The FAA/FAC protein complex was also not detected in FA cells from groups F, G, and H (data not shown). These results suggest that binding of FAA and FAC may be regulated by other proteins in the cell, perhaps encoded by other FA genes.
Figure 3.
Absence of FAA/FAC binding in cells derived from FA-B and FA-E patients. The indicated lymphoblast lines were analyzed for expression and binding of the FAA and FAC proteins. Protein from the indicated cell lines was immunoprecipitated (IP) with the anti-FAA (C-terminal) antiserum. Alternatively, proteins from an extract of a wt cell line were immunoprecipitated with a preimmune antiserum (Pre), where indicated. Protein was electrophoresed, transferred to nitrocellulose, and immunoblotted with either anti-FAA(N) or an anti-FAC antiserum. A whole-cell extract (WCE) from each cell line was analyzed in parallel for endogenous FAC expression (Bottom). (A) The cell lines examined were wt, FA-A (HSC72, lane 4 and PD9, lane 5), FA-B (HSC230), and FA-E (EUFA130), as indicated. (B) Cell lines were wt PD7, FA-A (HSC72), and FA-D (PD20), as indicated.
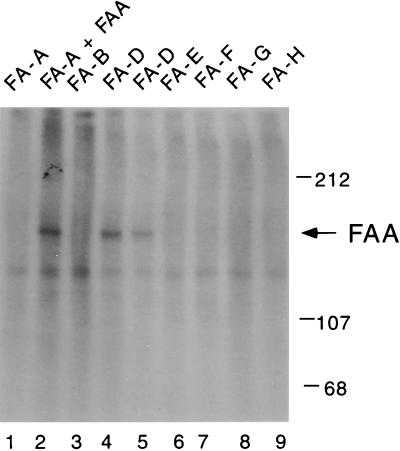
Various FA cell lines were next examined for FAA phosphorylation. Interestingly, the FAA protein was not phosphorylated in FA cells derived from groups A, B, C, E, F, G, and H (Fig. 4), but was phosphorylated in FA-D cells. These results show that FAA phosphorylation correlates with FAA/FAC binding.
Figure 4.
The FAA protein is not phosphorylated in groups B and E–H cell lines. The indicated lymphoblast lines were labeled with [32P]orthophosphate, and the FAA protein was immunoprecipitated with an affinity-purified anti-FAA (C-terminal) antiserum. Lymphoblast lines included HSC72 (lane 1), HSC72/pMMP-FAA (lane 2), HSC230 cells (lane 3), HSC62 cells (lane 4), PD20 cells (lane 5), EUFA130 cells (lane 6), EUFA121 (lane 7), EUFA143 (lane 8), and EUFA173 (lane 9).
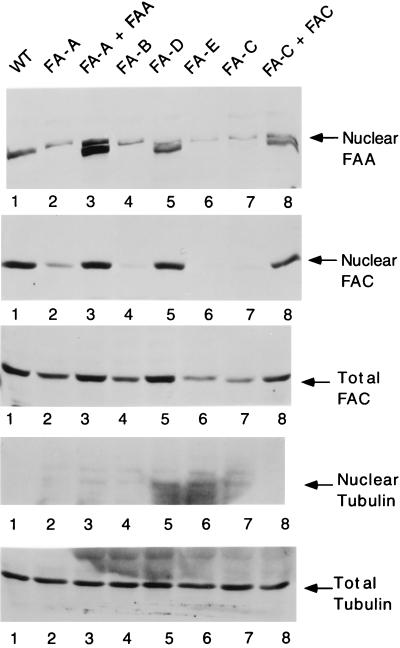
Previous studies have shown that FAC is expressed mostly in the cytoplasm (21, 28) with some nuclear expression as well (29). Although FAA has a nuclear localization signal, overexpressed FAA was found predominantly in the cytoplasm in one study (30). A complex of FAA and FAC protein was observed in both cytoplasmic and nuclear compartments (11, 31). To help resolve these conflicting studies, we next tested the localization of FAA and FAC in the various FA complementation groups (Fig. 5). All cell lines expressed relatively equal levels of FAA protein (Figs. 1 and 3). FAA protein was found in the nuclear fraction of normal lymphoblasts (Fig. 5, lane 1) and FA-D lymphoblasts (Fig. 5, lane 5) but was not found in the nuclear fraction of cells derived from groups A, B, E, and C (Fig. 5, lanes 2, 4, 6, and 7, respectively). All cell lines expressed relatively equal levels of FAC (Fig. 5, Total FAC immunoblot). Although FAC was observed in the nuclear fractions of all cell lines, nuclear FAC levels were increased in normal lymphoblasts, functionally complemented lymphoblasts, and FA-D lymphoblasts (Fig. 5, Nuclear FAC immunoblot, lanes 1, 3, 5, and 8). Taken together, these results show that FA cells derived from groups A, B, C, and E are defective in FAA/FAC binding (Fig. 3) and nuclear accumulation (Fig. 5). The data further support the functional correlation between formation and nuclear accumulation of the FAA/FAC complex.
Figure 5.
Absence of FAA and FAC in the nucleus of FA cells derived from complementation groups A, B, C, and E. The indicated lymphoblast lines were fractionated into total, cytoplasmic, and nuclear extracts as described in Lymphoblast Fractionation. Fractions were analyzed by immunoblot with either anti-FAA(N), anti-FAC(C), or anti-β-tubulin antibody. β-tubulin was excluded from the nuclear fractions, indicating proper cell fractionation. Cell lines tested included PD7 (lane 1), EUFA689 (lane 2), EUFA689/pMMP-FAA (lane 3), HSC230 (lane 4), PD20 (lane 5), EUFA130 (lane 6), HSC536 (lane 7), and HSC536/pMMP-FAC (lane 8). For the anti-FAA immunoblot, the FAA protein is the lower band of the doublet (lane 8) as described (11).
DISCUSSION
Several conclusions regarding the biochemical events in the FA signaling pathway are possible. First, binding of FAA and FAC proteins correlates with the biological function of the FA proteins (Table 1). In FA-A cell lines expressing mutant FAA protein FAAdelF1263 no FAA/FAC binding was observed. FAA/FAC binding was also not observed in FA-B and FA-E cells (Fig. 3) or in FA-F, FA-G, or FA-H cells (data not shown). These results suggest that the products of other FA genes regulate or stabilize the FAA/FAC protein complex. Consistent with this hypothesis, in vitro translated FAA and FAC proteins fail to bind directly (A.D. and I.G.-H., unpublished results), and other cellular factors may be required to assemble the FAA/FAC complex.
Table 1.
Summary of FA protein functional assays
| Cell type | FAA protein expressed | FAC protein expressed | FAA/FAC binding | FAA phosphorylated | FAA/FAC nuclear localization | MMC resistant |
|---|---|---|---|---|---|---|
| WT | + | + | + | + | + | + |
| FA-A | delF1263 | + | − | − | − | − |
| FA-B | + | + | − | − | − | − |
| FA-C | + | L554P | − | − | − | − |
| FA-D | + | + | + | + | + | − |
| FA-E | + | + | − | − | − | − |
| FA-F | + | + | − | − | ND | − |
| FA-G | + | + | − | − | ND | − |
| FA-H | + | + | − | − | ND | − |
ND, not determined.
Second, the phosphorylation of the FAA protein correlates with signaling through the FA pathway (Table 1). Mutations in the FAA and FAC protein that ablate protein binding also disrupt FAA phosphorylation. FAA phosphorylation was not observed in multiple FA complementation groups, suggesting that other FA genes regulate this signaling event. Several models may account for these observations. The interaction of FAA and FAC may be required for the subsequent binding and/or action of a protein kinase. Though our previous studies show that FAC interacts with the cyclin-dependent kinase, cdc2 (14), the kinase(s) required for FAA phosphorylation is currently unknown. Alternatively, FAA phosphorylation may be required for FAC binding to occur. For example, a phosphorylated amino acid residue of FAA may promote binding with FAC or may allow translocation of FAA to a cellular compartment in which FAC binding occurs. Without FAC binding, phosphorylated FAA may be rapidly dephosphorylated or degraded. Mapping of the FAA/FAC binding sites and FAA phosphorylation sites and identification of the relevant FAA protein kinase(s) would help to distinguish these models.
Third, the nuclear localization of the FAA/FAC protein complex correlates with function of the FA pathway (Table 1). In the current study, FAA and FAC localized to the nucleus in wt cells, functionally complemented cells, and FA-D cells. Interestingly, the nuclear complex is only weakly detected or absent in cells from other FA complementation groups, including groups A, B, C, and E. Moreover, the FA proteins affect the accumulation of the protein complex in the nucleus. FAA promotes the nuclear accumulation of FAC (11), and FAC promotes the nuclear accumulation of FAA (Fig. 5).
The molecular basis of the FAAdelF1263 mutation remains unclear, and several models are possible. The mutation may directly disrupt the FAC binding domain of the FAA protein or may prevent the normal compartmentalization of the FAA protein, thereby indirectly preventing FAC binding. Alternatively, the mutation may disrupt kinase binding and/or FAA phosphorylation. Interestingly, FAAdelF1263 seems to be a relatively prevalent mutation of FAA, observed in 30 patients with FA-A (26). Other less common point mutations also map to an adjacent region of the FAA gene (26, 27), suggesting that this region, containing exons 32–39 (32), may encode a critical functional domain of the FAA protein. Further studies are required to determine whether the FAAdelF1263 mutation or other patient-derived point mutations directly block the ability of FAA to correct the MMC sensitivity of a heterologous FA-A cell line.
The temporal sequence of events in the FA pathway also will require further testing. Because the FAA/FAC complex is present in both cytoplasmic and nuclear compartments (11), there are at least three possibile sequences: (i) phosphorylation, binding, then nuclear translocation, (ii) binding, nuclear translocation, then phosphorylation, and (iii) binding, phosphorylation, then nuclear translocation. Determining the localization of the phosphorylated form of FAA may help to identify the actual sequence.
Interestingly, FA-D is distinct from other FA complementation groups. FA-D cells are sensitive to MMC (23), despite normal levels of FAA and FAC proteins, FAA/FAC binding, FAA phosphorylation, and normal nuclear accumulation of the FAA/FAC complex (Table 1). This unique phenotype has been confirmed in cell lines from four independent patients with FA-D (data not shown). These observations suggest that the FA-D gene product functions downstream or independently of the FA protein complex, perhaps through its recruitment to the FA pathway in the nucleus. It is not yet known whether the four patients with FA-D differ significantly from other patients with FA with respect to their clinical phenotype or cancer susceptibility.
Finally, the identification of additional protein components of the FAA/FAC complex may help to define the biochemical function(s) of the complex and may allow the identification of other FA-gene products. For instance, recent studies have shown that the tumor suppressor protein BRCA1 interacts with the DNA repair protein Rad51 (33, 34). Other protein complexes, such as the Rad50/Mre11/nibrin complex are defective in known chromosome-instability syndromes (35, 36). It remains to be determined whether the FA proteins interact similarly with known DNA repair proteins in the nucleus.
Acknowledgments
We thank Markus Grompe for PD20, PD9, and PD475 cell lines and R. Mulligan for the pMMP-retroviral vector and the 293GPG producer cells. This work was supported by National Institutes of Health Grants K08-H103420 (to G.M.K.) and R01-HL52725 and PO1-HL54785 (to A.D.D.). D.N. is a Fellow and A.D.D. is a Scholar of the Leukemia Society of America.
ABBREVIATIONS
- FA
Fanconi anemia
- MMC
mitomycin C
- wt
wild type
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1. D’Andrea A D, Grompe M. Blood. 1997;90:1725–1736. [PubMed] [Google Scholar]
- 2.Liu J, Buchwald M, Walsh C E, Young N S. Blood. 1994;84:3995–4007. [PubMed] [Google Scholar]
- 3.Auerbach A, Buchwald M, Joenje H. In: The Metabolic and Molecular Bases of Inherited Disease [CD-ROM] Scriver A, Beaudet L, Sly W S, Valle D, editors. New York: McGraw–Hill; 1997. [Google Scholar]
- 4.Strathdee C A, Duncan A M V, Buchwald M. Nat Genet. 1992;1:196–198. doi: 10.1038/ng0692-196. [DOI] [PubMed] [Google Scholar]
- 5.Joenje H, Ten F L, Oostra A, Berkel C V, Rooimans M, Schroeder S, Kurth T, Wegner R, Gille J, Buchwald M, Arwert F. Blood. 1995;86:2156–2160. [PubMed] [Google Scholar]
- 6.Joenje H, Oostra A B, Wijker M, di Summa F M, van Berkel C G M, Rooimans M A, Ebell W, van Weel M, Pronk J C, Buchwald M, Arwert F. Am J Hum Genet. 1997;61:940–944. doi: 10.1086/514881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Strathdee C A, Gavish H, Shannon W R, Buchwald M. Nature (London) 1992;356:763–767. doi: 10.1038/356763a0. [DOI] [PubMed] [Google Scholar]
- 8.Lo Ten Foe J R, Rooimans M A, Bosnoyan-Collins L, Alon N, Wijker M, Parker L, Lightfoot J, Carreau M, Callen D F, Savoia A, et al. Nat Genet. 1996;14:320–323. doi: 10.1038/ng1196-320. [DOI] [PubMed] [Google Scholar]
- 9.The Fanconi Anemia/Breast Cancer Consortium. Nat Genet. 1996;14:324–328. [Google Scholar]
- 10.Buchwald M. Nat Genet. 1995;11:228–230. doi: 10.1038/ng1195-228. [DOI] [PubMed] [Google Scholar]
- 11.Kupfer G M, Naf D, Suliman A, Pulsipher M, D’Andrea A D. Nat Genet. 1997;17:487–490. doi: 10.1038/ng1297-487. [DOI] [PubMed] [Google Scholar]
- 12.Kaiser T N, Lojewski A, Dougherty C, Juergens L, Sahar E, Latt S A. Cytometry. 1982;2:291–297. doi: 10.1002/cyto.990020505. [DOI] [PubMed] [Google Scholar]
- 13.Kubbies M, Schindler D, Hoehn H, Schinzel A, Rabinovich P S. Am J Hum Genet. 1985;37:1022–1030. [PMC free article] [PubMed] [Google Scholar]
- 14.Kupfer G, Yamashita T, Naf D, Suliman A, Asano S, D’Andrea A D. Blood. 1997;90:1047–1054. [PubMed] [Google Scholar]
- 15.Rathbun R, Faulkner G, Ostroski M, Christianson T, Hughes G, Jones G, Cahn R, Maziarz R, Royle G, Keeble W, et al. Blood. 1997;90:974–985. [PubMed] [Google Scholar]
- 16.Whitney M A, Royle G, Low M J, Kelly M A, Axthelm M K, Reifsteck C, Olson S, Braun R E, Heinrich M C, Rathbun R K, et al. Blood. 1996;88:49–58. [PubMed] [Google Scholar]
- 17.Cumming R C, Liu J M, Youssoufian H, Buchwald M. Blood. 1996;88:4558–4567. [PubMed] [Google Scholar]
- 18.Marathi U K, Howell S R, Ashmun R A, Brent T P. Blood. 1996;88:2298–2305. [PubMed] [Google Scholar]
- 19.Ridet A, Guillouf C, Duchaud E, Cundari E, Fiore M, Moustacchi E, Rosselli F. Cancer Res. 1997;57:1722–1730. [PubMed] [Google Scholar]
- 20.Chen M, Tomkins D J, Auerbach W, McKerlie C, Youssoufian H, Liu L, Gan O, Carreau M, Auerbach A, Groves T, et al. Nat Genet. 1996;12:448–451. doi: 10.1038/ng0496-448. [DOI] [PubMed] [Google Scholar]
- 21.Yamashita T, Barber D L, Zhu Y, Wu N, D’Andrea A D. Proc Natl Acad Sci USA. 1994;91:6712–6716. doi: 10.1073/pnas.91.14.6712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamashita T, Wu N, Kupfer G, Corless C, Joenje H, Grompe M, D’Andrea A D. Blood. 1996;87:4424–4432. [PubMed] [Google Scholar]
- 23.Whitney M, Thayer M, Reifsteck C, Olson S, Smith L, Jakobs P M, Leach R, Naylor S, Joenje H, Grompe M. Nat Genet. 1995;11:341–343. doi: 10.1038/ng1195-341. [DOI] [PubMed] [Google Scholar]
- 24.Pulsipher M, Kupfer G M, Naf D, Suliman A, Lee J-S, Jakobs P, Grompe M, Joenje H, Sieff C, Guinan E, et al. Mol Med. 1998;4:468–479. [PMC free article] [PubMed] [Google Scholar]
- 25.Ory D, Neugeboren B, Mulligan R. Proc Natl Acad Sci USA. 1996;93:11400–11406. doi: 10.1073/pnas.93.21.11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levran O, Erlich T, Magdalena N, Gregory J J, Batish S D, Verlander P C, Auerbach A D. Proc Natl Acad Sci USA. 1997;94:13051–13056. doi: 10.1073/pnas.94.24.13051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wijker, M., Morgan, N. V., Herterich, S., van Berkel, C. G. M., Tipping, A. J., Gross, H. J., Gille, J. J. P., Pals, G., Savino, M., Altay, C., et al. (1998) Eur. J. Hum. Genet., in press. [DOI] [PubMed]
- 28.Youssoufian H. Proc Natl Acad Sci USA. 1994;91:7975–7979. doi: 10.1073/pnas.91.17.7975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoatlin M E, Christianson T A, Keeble W W, Hammond A T, Zhi Y, Heinrich M C, Tower P A, Bagby G C., Jr Blood. 1998;91:1418–1425. [PubMed] [Google Scholar]
- 30.Kruyt F A E, Waisfisz Q, Dijkmans L M, Hermsen M A, Youssoufian H, Arwert F, Joenje H. Blood. 1997;90:3288–3295. [PubMed] [Google Scholar]
- 31.Naf D, Kupfer G M, Suliman A, Lambert K, D’Andrea A D. Mol Cell Biol. 1998;18:5952–5960. doi: 10.1128/mcb.18.10.5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ianzano L, D’Apolito M, Centra M, Savino M, Levran O, Auerbach A D, Cleton-Jansen A-M, Doggett N A, Pronk J C, Tipping A J, et al. Genomics. 1997;41:309–314. doi: 10.1006/geno.1997.4675. [DOI] [PubMed] [Google Scholar]
- 33.Scully R, Chen J, Plug A, Xiao Y, Weaver D, Feunteun J, Ashley T, Livingston D M. Cell. 1997;88:265–275. doi: 10.1016/s0092-8674(00)81847-4. [DOI] [PubMed] [Google Scholar]
- 34.Scully R, Chen J, Ochs R L, Keegan K, Hoekstra M, Feunteun J, Livingston D M. Cell. 1997;90:425–435. doi: 10.1016/s0092-8674(00)80503-6. [DOI] [PubMed] [Google Scholar]
- 35.Varon R, Vissinga C, Platzer M, Cerosaletti K M, Chrzanowska K H, Saar K, Beckmann G, Seemanova E, Cooper P R, Nowak N J, et al. Cell. 1998;93:467–476. doi: 10.1016/s0092-8674(00)81174-5. [DOI] [PubMed] [Google Scholar]
- 36.Carney J P, Maser R S, Olivares H, Davis E M, Le Beau M, Yates J R, III, Hays L, Morgan W F, Petrini J H J. Cell. 1998;93:477–486. doi: 10.1016/s0092-8674(00)81175-7. [DOI] [PubMed] [Google Scholar]
- 37.Verlander P C, Kaporis A, Liu Q, Zhang Q, Seligsohn U, Auerbach A D. Blood. 1995;86:4034–4038. [PubMed] [Google Scholar]