Abstract
Low caloric intake (caloric restriction) can lengthen the life span of a wide range of animals and possibly even of humans. To understand better how caloric restriction lengthens life span, we used genetic methods and criteria to investigate its mechanism of action in the nematode Caenorhabditis elegans. Mutations in many genes (eat genes) result in partial starvation of the worm by disrupting the function of the pharynx, the feeding organ. We found that most eat mutations significantly lengthen life span (by up to 50%). In C. elegans, mutations in a number of other genes that can extend life span have been found. Two genetically distinct mechanisms of life span extension are known: a mechanism involving genes that regulate dauer formation (age-1, daf-2, daf-16, and daf-28) and a mechanism involving genes that affect the rate of development and behavior (clk-1, clk-2, clk-3, and gro-1). We find that the long life of eat-2 mutants does not require the activity of DAF-16 and that eat-2; daf-2 double mutants live even longer than extremely long-lived daf-2 mutants. These findings demonstrate that food restriction lengthens life span by a mechanism distinct from that of dauer-formation mutants. In contrast, we find that food restriction does not further increase the life span of long-lived clk-1 mutants, suggesting that clk-1 and caloric restriction affect similar processes.
It was shown more than 50 years ago that reducing the caloric intake (caloric restriction) of rodents can significantly lengthen their mean and maximal life span (1). It subsequently has been shown that caloric restriction (CR) can lengthen the life span of a wide variety of animals (2). Trials have even begun with higher primates; based on preliminary evidence, calorically restricted rhesus monkeys show similar signs of delayed aging to those seen in the calorically restricted rodents (3–7). CR has been best studied in rodents, and it is known that rodents undergoing CR display many physiological changes, including reduced body weight, temperature, blood glucose, and insulin levels (reviewed in refs. 8 and 9). However, it is unclear which of these changes are required for an extended life span (8, 9). Several studies indicate that reducing caloric intake reduces the amount of damage attributable to free radicals (reviewed in refs. 8 and 9). One simple hypothesis to explain how CR extends life span is that CR may reduce basal metabolic rates. Rodents and primates undergoing CR have lowered body temperatures (10–12), an indication of lower metabolic rates. However, studies on the effect of CR on the metabolic rates of various mammals have given equivocal results, with some studies showing no change in oxygen consumption per unit of lean body mass (13, 14) and other studies showing a decrease of consumption under CR (15, 16). In spite of uncertainty about how CR affects life span, it remains the only experimental treatment that has been shown repeatedly to significantly prolong the life of vertebrates (8, 9, 17).
On the other hand, it is in Caenorhabditis elegans that genetic mechanisms that can extend life span have been best characterized, and the worm has become the foremost animal model system for the molecular analysis of life span (18–20). At least two different genetic mechanisms for extending life span have been identified in C. elegans. One mechanism involves the partial activation of the dauer pathway by mutations in the genes age-1, daf-2, and daf-28 (reviewed in refs. 18–20). age-1 and daf-2 are involved in an insulin-like signaling cascade that regulates the activity of the forkhead-like transcription factor DAF-16 (21–24). Loss-of-function mutations in daf-16 strongly suppress the extremely long life of the dauer mutants age-1 and daf-2, as well as all other phenotypes of daf-2 and age-1 (23–28). The second mechanism involves the reduction of physiological rates by mutations in the Clock genes clk-1, clk-2, clk-3, and gro-1 (12, 29, 30). So far, only one of these genes, clk-1, has been cloned. clk-1 encodes a small protein of unknown biochemical function that may regulate metabolism (31).
Lowering food intake also lengthens C. elegans life span, indicating that the mechanism of CR may exist in the worm (32, 33). We wondered whether any of the known genetic factors extending life span involve the same mechanism as food restriction. To study food restriction directly, worms can be grown in liquid culture with different known concentrations of bacteria. However, most aging studies in the worm have been done under standard C. elegans culture conditions (i.e., worms are cultured on agar plates with lawns of Escherichia coli bacteria). Therefore, the response of many long-lived C. elegans strains to growth in liquid culture are unknown, and it is not even clear whether all of these strains can be cultured under such conditions. Doing aging studies in liquid culture also poses difficult technical problems such as accurately maintaining and reproducing certain bacterial concentrations. Consequently, we wanted to study food restriction under standard experimental conditions to compare our results directly to previously published work on the genetic basis of life span in C. elegans. To achieve this, we used eat mutations to reduce the food intake of worms.
eat mutants have defects in pharyngeal function leading to an insufficient food uptake and a starved appearance (34). In wild-type worms, the pharyngeal muscles contract to ingest bacteria more than 200 times per minute (35). Some eat mutations affect the strength or the proper sequence of contractions and relaxations of the various muscles of the pharynx, leading to inefficient feeding (34). Mutations in other genes, however, do not affect the coordination of the muscle contraction, but only drastically reduce the rate of pharyngeal pumping (34, 35). eat mutations were identified based on visible defects in feeding behavior that resemble those caused by experimental alterations in the muscles or nervous system of the pharynx (34). For some eat mutants, there is pharmacological, electrophysiological, genetic, or molecular evidence that they affect the nervous system or muscles (34–38).
In this paper we show that many eat mutations prolong life span, almost certainly by restricting the caloric intake of the worm. We also demonstrate that food restriction lengthens the life span of C. elegans by a mechanism that is genetically distinct from that induced by mutations in the dauer genes, but which is similar to that induced by mutations in the gene clk-1.
MATERIALS AND METHODS
Aging Experiments.
Aging experiments were performed as described (30) except that experiments were begun by allowing adult hermaphrodites to lay eggs for a limited time (4–6 hr). Animals were cultured at 20°C unless otherwise stated. Worms were examined every day until death and were scored as dead when they were no longer able to move even in response to prodding with a platinum pick. Each day, any dead worms were removed from plates and the deaths were recorded. Experiments were started with 50 experimental worms per genotype (10 per plate) and the wild type (N2) was always included as a control. A plate of approximately 30 spare worms was started at the same time as the experimental worms and was treated identically, except that deaths on this plate were not counted. Worms that died from matricidal hatching (the bag-of-worms phenotype) were replaced by spare worms when possible or were discarded from the analysis when spare worms were exhausted.
Strains Used.
We used eat mutations to reduce the food intake of worms while they were maintained under standard experimental conditions. By maintaining such conditions, we can directly compare our results with previously published work on the genetic basis of life span in C. elegans. All eat mutants available from the Caenorhabditis Genetics Center were examined except those with mutations in eat-9, -11, -14, -16 and egl-19 ( = eat-12). Three other alleles of eat-2 and two alleles of eat-18, kindly provided by L. Avery and D. Raisen (University of Texas Southwestern Medical Center), also were examined. A selection of uncoordinated (unc) mutations also were examined. These mutations were chosen to represent a wide cross section of primary defects. However, unc mutations that lead to an egg laying-defective (Egl) phenotype were explicitly excluded, because most of these mutants die from matricidal hatching.
New Alleles of unc-79 and unc-80.
Mutations in unc-79 and unc-80 cause subtle movement defects. New alleles of unc-79 and unc-80 were recovered in a screen for maternal-effect viable mutations (39). These mutations subsequently were found to be fully zygotic mutations that fail to complement either unc-79(e1030) or unc-80(e1272).
Backcrossing eat and unc Strains.
In a preliminary study, we found that some strains containing unc mutations and some strains containing eat mutations could live longer than the wild type, whereas others did not (unpublished results). As the pattern of mutations that extended life span was difficult to interpret, we suspected that background effects could be affecting the life span of some strains. Many C. elegans strains available from the Caenorhabditis Genetics Center have been backcrossed only once or twice to the wild type and are likely to carry many background mutations. As a precaution against background effects, all of the eat and many of the unc mutations were backcrossed twice to the wild type before measuring life span. Because no significant differences in the effect of eat mutations on life span before and after backcrosses were noted (unpublished data), the strain DA465 eat-2(ad465) was used without backcrosses in experiments testing the interaction of eat-2 with daf-16, daf-2, and clk-1.
Strain Construction.
Double-mutant strains were constructed by using standard techniques (40, 41) selecting for easily identifiable phenotypes: slow growth (clk-1), Daf-c (daf-2), Eat (eat-2), and the ability to suppress the Daf-c phenotype of daf-2 (daf-16).
Statistical Analysis.
Statistics were calculated by using the Microsoft excel 97 analysis ToolPak. Mean life spans were compared by using Student’s t test assuming unequal variances.
RESULTS
Most eat Mutations Lengthen Life Span.
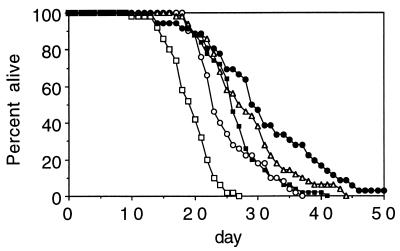
We tested the life span of a large number of eat mutations to see whether these mutations extend life span. As expected, we found that mutations in many eat genes (eat-1, eat-2, eat-3, eat-6, eat-13, and eat-18) significantly extend mean and maximum life span (Table 1). All four tested alleles of eat-2 and eat-6, as well as both alleles of eat-1 and eat-18, significantly increase life span (Fig. 1, Table 1). Of all of the eat genes tested, the strongest effect was seen with eat-2 mutants, which can live over 50% longer than the wild type (Table 1; Fig. 1). The life span extension of eat-2 is comparable to other previously characterized long-lived mutants, such as clk-1(e2519) and daf-2(e1370) (ref. 30; Fig. 3), and is of similar magnitude to the effect of CR on the life span of mammals (42). The effect of eat-2 on life span is also highly reproducible. The life span of eat-2(ad465) has been determined seven times, and in every trial mutants live substantially longer than the wild type, with an average lengthening of life span of 47% over the wild type (Table 1; Fig. 1 and 2B; and data not shown).
Table 1.
Mean life span of a number of eat mutant strains
| Strain | Genotype | n | Mean | Diff, % | Significance level |
|---|---|---|---|---|---|
| Experiment no. 1 | |||||
| N2 | + | 50 | 21.6 ± 0.6 | — | — |
| MQ581 | eat-1(ad427) | 50 | 28.8 ± 1.0 | +33 | <0.0005 |
| MQ582* | eat-1(e2343) | 50 | 23.9 ± 0.9 | +11 | 0.03 |
| MQ573 | eat-5(ad464) | 50 | 20.2 ± 0.5 | −6 | 0.07 |
| MQ574 | eat-7(ad450) | 50 | 14.0 ± 0.5 | −35 | <0.0005 |
| Experiment no. 2 | |||||
| N2 | + | 50 | 19.5 ± 0.5 | — | — |
| MQ643 | eat-2(ad453) | 50 | 26.5 ± 0.7 | +36 | <0.0005 |
| MQ594 | eat-2(ad465) | 50 | 25.1 ± 0.7 | +29 | <0.0005 |
| MQ644 | eat-2(ad1113) | 50 | 28.4 ± 0.9 | +46 | <0.0005 |
| MQ631 | eat-2(ad1116) | 36 | 30.6 ± 1.4 | +57 | <0.0005 |
| MQ646 | eat-18(ad820sd) | 50 | 26.9 ± 1.0 | +38 | <0.0005 |
| MQ649 | eat-18(ad1110) | 38 | 22.4 ± 0.9 | +15 | 0.008 |
| Experiment no. 3 | |||||
| N2 | + | 50 | 19.9 ± 0.5 | — | — |
| MQ676 | eat-3(ad426) | 50 | 22.0 ± 0.9 | +11 | 0.04 |
| MQ584 | eat-6(ad792) | 50 | 27.1 ± 0.6 | +36 | <0.0005 |
| MQ583 | eat-6(ad467) | 50 | 27.2 ± 0.7 | +37 | <0.0005 |
| MQ591 | eat-6(ad601) | 50 | 22.8 ± 0.7 | +15 | 0.001 |
| MQ645 | eat-6(ad997) | 39 | 23.6 ± 0.7 | +19 | <0.0005 |
| MQ677 | eat-10(ad606) | 31 | 21.5 ± 0.8 | +8 | 0.12 |
| MQ596 | eat-13(ad522) | 50 | 26.0 ± 0.6 | +31 | <0.0005 |
Sample size (n), mean life span ± SEM (in days), and percent difference from the wild type are given. The final column displays the probability that the mean life span is the same as the wild type (Student’s t test). Strains that lived significantly longer than the wild type (P < 0.05) are shown in bold, and short-lived strains are underlined. Three separate experiments were performed at different times, each with the wild type (N2) as a control.
A high proportion of eat-1(e2343) worms died from a prolapsed gonad. If these worms are subtracted from the analysis, the mean life span increases significantly. We also tested the life span of eat-8(ad599), eat-15(ad602), and eat-17(ad707); however, the sample sizes for these strains were too low to give reliable results because of high levels of matricidal hatching. eat-4(ad572), eat-4(ad819), and eat-4(ky5) also were tested, but most of the worms crawled off the plates and died from desiccation. The life span of DA531 eat-1(ad427), DA631 eat-3(ad426); him-8(e1489), DA464 eat-5(ad464), DA467 eat-6(ad467), and DA521 eat-7(ad450) were determined twice, and that of DA465 eat-2(ad465) was determined six times. The life span of these strains was very similar to respective backcrossed strains presented above.
Figure 1.
Four alleles of eat-2 lengthen life span. The percentage of worms alive on a given day after eggs being laid for a single experiment: N2 (□), eat-2(ad465) (○), eat-2(ad453) (■), eat-2(ad1113) (▵), and eat-2(ad1116) (•). Mean life spans are given in Table 1. The death of the last surviving eat-2(ad1116) worm is not shown. This worm died on day 86.
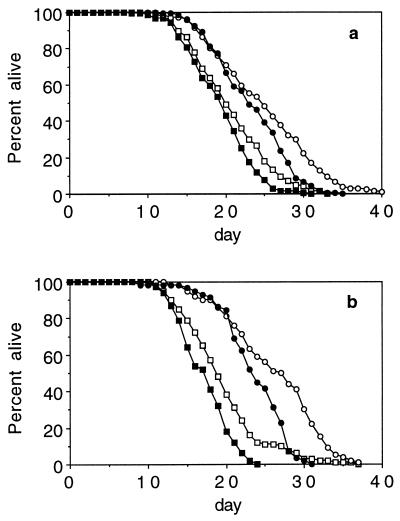
Figure 3.
The interaction of eat-2 with daf-2 and clk-1. (a) The percentage of worms alive on a given day after eggs being laid (day 0) at 20°C: N2(□), eat-2(ad465)(■), daf-2(e1370) (▴) and MQ413 eat-2(ad465); daf-2(e1370) (•). Mean life span ± SEM, with sample size in parentheses, are 21.9 ± 0.8 (50), 26.3 ± 0.9 (50), 34.1 ± 1.6 (66), and 41.6 ± 2.1 (60), respectively. eat-2(ad465); daf-2(e1370) worms live significantly longer than either eat-2(ad465) or daf-2(e1370) (P < 0.0005 and P = 0.005, respectively). A second trial with these strains gave very similar results. (b) The percentage of worms alive on a given day after eggs being laid (day 0) for two pooled experiments at 20°C: N2 (□), eat-2(ad465) (■), clk-1(e2519) (•) and eat-2(ad465); clk-1(e2519) (○). Mean life spans are 19.7 ± 0.5 (100), 26.3 ± 0.6 (100), 25.1 ± 0.9 (100), and 27.5 ± 0.8 (100), respectively. eat-2(ad465); clk-1(e2519) lives only marginally longer than clk-1(e2519) (P = 0.05) and no longer than eat-2(ad465) (P = 0.23). In one trial, clk-1(qm30) and eat-2(ad465); clk-1(qm30) were scored. The results for this experiment were: N2, 21.9 ± 0.8 (50); eat-2, 26.3 ± 0.9 (50); clk-1, 24.1 ± 1.4 (50), and eat-2; clk-1, 26.5 ± 1.5 (50).
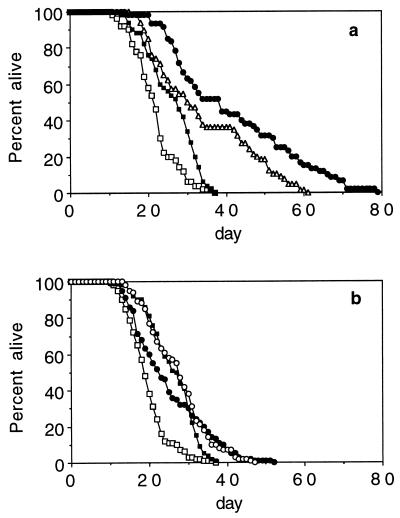
Figure 2.
The interaction of daf-16 with clk-1 and eat-2. (a) The percentage of worms alive on a given day after eggs being laid (day 0) for three pooled experiments at 18°C: N2(□), daf-16(m26)(■), clk-1(e2519) (○), and daf-16(m26); clk-1(e2519) (•). Mean life span ± SEM, with sample size in parentheses, are 20.8 ± 0.4 (145), 19.0 ± 0.6 (145), 25.0 ± 0.5 (141) and 23.5 ± 0.4 (149), respectively. (b) The percentage of worms alive on a given day after eggs being laid (day 0) for two pooled experiments at 20°C: N2 (□), daf-16(m26) (■), eat-2(ad465) (○), and daf-16(m26); eat-2(ad465) (•). Mean life spans are 19.7 ± 0.5 (100), 17.4 ± 0.3 (100), 26.3 ± 0.6 (100), and 23.6 ± 0.6 (57), respectively. In one of the two trials pooled for this figure, and in Fig. 3, the last few N2 worms lived much longer than normal. In this trial, the maximum life span of eat-2(ad465) and N2 were comparable. However, in the six other trials eat-2(ad465) had a maximum life span that was clearly greater than that of N2. All other eat mutations that lengthen mean life span also clearly lengthen maximum life span.
Three of the eat mutations tested did not lengthen life span. The mutations eat-5(ad464) and eat-10(ad606) do not affect life span (Table 1). It is not clear why these mutations do not lengthen life span, but it is possible that mutations in these genes may lead to a feeding defect that is too weak to affect life span. Alternatively, these mutations may produce deleterious pleiotropic effects that mask any positive effects of food restriction on life span. Surprisingly, the only known allele of another gene, eat-7, actually shortens life span (Table 1). However, the significance of this result is unclear, because eat-7 has a very unusual phenotype (34). eat-7(ad450) is a poorly studied semidominant mutation that causes worms to “fall asleep” when left undisturbed. In this state, worms do not feed, move, or defecate.
Life Span Correlates with the Severity of the Feeding Defect.
Thus, 14 of 17 eat mutations tested significantly lengthen life span, strongly suggesting that most eat mutants live long as a result of food restriction. This hypothesis is strengthened by the observation that in both eat-2 and eat-6 mutants, the severity of the eating defect appears to correlate with life span. Indeed, eat-6(ad792) and eat-6(ad467) have more severe feeding defects (36) and also live longer than eat-6(ad601) and eat-6(ad997). The case is even clearer for eat-2 mutants. Mutations in eat-2 appear to affect only the rate of pharyngeal pumping (35). As Klass (32) has shown that food intake is linearly related to pumping rate, pumping rate is a reliable measure of nutritional status. We find that the eat-2 alleles with the slowest pumping rates, eat-2(ad1113) and eat-2(ad1116) (35), live longer than one of the weakest eat-2 alleles, ad453 (Fig. 1; Table 1). In this experiment, another strong allele, ad465, did not lengthen life span as much as ad453 (29% versus 36% increase over the wild type). However, the life span of ad465 in this trial was much shorter than its average life span for seven trials (a 47% increase in life span over the wild type).
Most unc Mutations Do Not Lengthen Life Span.
The fact that long-lived eat mutants display a range of defects with different underlying molecular or anatomical causes strongly suggests that these mutants live long because of the only phenotype they share: restricted food intake. It remains possible, however, that the eat mutants live long not because of food restriction, but rather as a consequence of some pleiotropic effect on the nervous system and/or muscles. To test this possibility, we examined the life span of a number of unc mutants. unc mutants display movement defects caused by abnormalities in the function or development of the nervous system and/or the body-wall muscles (43). Thus, similar molecular defects may underlie the Eat and Unc phenotypes. This idea is borne out by the observation that eat-8 and eat-11 are also Unc, whereas unc-2, -10, -11, -17, -18, -26, -32, -36, -37, -57, -75, and -104 mutants are also Eat (34). Thus, unc mutations that do not affect pharyngeal pumping are excellent controls for the effect of eat mutations on life span.
We tested the effect of mutations in 15 unc genes on life span and found that mutations in 14 of these genes (unc-1, -4, -6, -7, -9, -24, -25, -29, -30, -46, -47, -49, -79, and -80) do not lengthen life span (Table 2 and data not shown). However, some unc strains appear to contain background mutations that can extend life span. The strains CB138 unc-24(e138) and CB927 unc-24(e927) contain background mutations that are not strongly linked to unc-24; they could be removed by outcrossing the unc-24 alleles to the wild type twice (Table 2). Three unc strains [containing the mutations unc-25(e156), unc-79(e1069), and unc-80(e1272)] contain life-extending background mutations that appear to be linked to the unc mutation; they could not be removed by two backcrosses (Table 2). However, other alleles of these genes do not affect life span, indicating that these genes do not affect life span (Table 2). Although the life span of unc mutants has not been systematically studied, some previous work has shown that at least five other unc genes (unc-2, 15, 20, 54, and 78) do not lengthen life span (44).
Table 2.
Mean life span of a number of unc mutant strains
| Strain | Genotype | n | Mean | Diff, % | Significance level |
|---|---|---|---|---|---|
| Experiment no. 1* | |||||
| N2 | + | 50 | 19.0 ± 0.5 | — | — |
| MQ545 | unc-24(e138) | 50 | 19.2 ± 0.6 | +1 | 0.79 |
| MQ546 | unc-24(e448) | 50 | 19.1 ± 0.4 | +1 | 0.85 |
| MQ547 | unc-24(e927) | 50 | 19.0 ± 0.5 | 0 | 0.95 |
| MQ548 | unc-24(e1172) | 50 | 19.7 ± 0.4 | +4 | 0.25 |
| Experiment no. 2 | |||||
| N2 | + | 50 | 21.7 ± 0.6 | — | — |
| MQ613 | unc-25(e265) | 50 | 22.8 ± 1.1 | +5 | 0.42 |
| MQ612 | unc-25(e591) | 50 | 22.1 ± 0.9 | +2 | 0.71 |
| MQ611 | unc-25(e891) | 50 | 22.5 ± 1.0 | +4 | 0.49 |
| MQ633 | unc-49(e382) | 50 | 23.1 ± 0.6 | +7 | 0.11 |
| MQ603 | unc-79(e1068) | 50 | 24.1 ± 0.9 | +11 | 0.03 |
| MQ604 | unc-80(e1069) | 50 | 21.6 ± 0.6 | 0 | 0.95 |
| Experiment no. 3 | |||||
| N2 | + | 50 | 19.9 ± 0.5 | — | — |
| MQ587 | unc-25(e156) | 50 | 25.1 ± 0.7 | +26 | <0.0005 |
| MQ592 | unc-26(e205) | 50 | 28.7 ± 0.6 | +44 | <0.0005 |
| MQ647 | unc-26(e345) | 34 | 29.2 ± 0.8 | +47 | <0.0005 |
| MQ574 | unc-26(e1196) | 50 | 27.3 ± 0.7 | +37 | <0.0005 |
| MQ648 | unc-26(m2) | 39 | 26.8 ± 0.7 | +35 | <0.0005 |
| MQ632 | unc-30(e596) | 50 | 18.2 ± 0.5 | −9 | 0.02 |
| Experiment no. 4 | |||||
| N2 | + | 50 | 21.6 ± 0.6 | — | — |
| MQ575 | unc-30(e191) | 50 | 20.0 ± 0.6 | −7 | 0.06 |
| MQ588 | unc-30(e318) | 50 | 16.9 ± 0.5 | −22 | <0.0005 |
| MQ578 | unc-46(e177) | 50 | 21.5 ± 1.0 | 0 | 0.96 |
| MQ579 | unc-47(e367) | 50 | 20.5 ± 0.5 | −5 | 0.16 |
| Experiment no. 5 | |||||
| N2 | + | 50 | 19.9 ± 0.7 | — | — |
| MQ636 | unc-79(e1030) | 38 | 19.1 ± 0.7 | −4 | 0.43 |
| MQ635 | unc-79(qm12) | 50 | 19.7 ± 0.5 | −1 | 0.89 |
| MQ634 | unc-79(qm14) | 50 | 21.9 ± 0.8 | +10 | 0.05 |
| MQ638 | unc-80(e1272) | 50 | 22.0 ± 0.7 | +11 | 0.03 |
| MQ608 | unc-80(qm2) | 50 | 21.2 ± 0.7 | +7 | 0.18 |
| MQ609 | unc-80(qm3) | 50 | 20.1 ± 0.6 | +1 | 0.83 |
| MQ637 | unc-80(qm9) | 50 | 19.5 ± 0.6 | −2 | 0.67 |
Columns are as described for Table 1. Before outcrossing all of the strains listed above, a number of other unc strains received from the Caenorhabditis Genetics Center or Medical Research Council (United Kingdom) also were tested. From these studies we found that mutations in the genes unc-1, -4, -6, -7, -9, and -29 do not affect life span (unpublished data).
unc-24 alleles have been tested four times with similar results.
Of the 15 unc genes that we examined, only mutations in unc-26 clearly lengthen life span. After backcrossing four alleles of unc-26 to the wild type twice, we determined that all four alleles significantly lengthen life span. However, because unc-26 mutants are known to have a starved appearance and a feeding defect (34), the long life of these mutants is probably also the result of food restriction. Thus, mutations in 14 of 14 tested unc genes that do not affect pharyngeal pumping do not lengthen life span, whereas mutations in 7 of 10 genes that disrupt normal pharyngeal pumping do lengthen life span. This observation, along with the other evidence presented, indicates that eat genes indeed lengthen life span by reducing food intake and presumably caloric intake.
At Least Two Genetic Mechanisms Can Lengthen C. elegans Life Span.
We wondered whether any of the known genetic factors extending life span in C. elegans involve the same mechanism as food restriction. Previously, we provided evidence for the existence of at least two distinct genetic mechanisms in C. elegans that can extend life span. Specifically, we showed that the dauer genes (daf-2, age-1, and daf-16) are involved in a mechanism that is distinct from that in which the Clock genes (clk-1, clk-3, and gro-1) are involved (30). Our main evidence was that mutations in daf-16 that do suppress the long life of daf-2 and age-1 mutants do not suppress the long life of the Clock mutants (30). However, the result for clk-1 has been disputed (45). To solve the question of the independence of the two pathways, we have retested the life span of daf-16; clk-1 double mutants (Fig. 2A) by using the reference alleles of these genes. daf-16(m26) mutants live slightly shorter than the wild type (Fig. 2A), a result that we have seen repeatedly and that also has been noted by others (26, 27, 46). We find that daf-16(m26) has a similar, slightly deleterious effect on clk-1 mutants; that is, daf-16(m26); clk-1(e2519) double mutants live slightly shorter than clk-1(e2519) mutants. However, daf-16(m26); clk-1(e2519) double mutants still live significantly longer than the wild type (Fig. 2A). This observation indicates that the shorter life span of daf-16(m26); clk-1(e2519) as compared with clk-1(e2519) is not caused by partial suppression of the long life of clk-1(e2519) by daf-16(m26). Rather, this suggests that the daf-16(m26) mutation, or a mutation strongly linked to it, has a weak deleterious effect on life span. That the dauer genes age-1 and daf-2 lengthen life span by a different mechanism than clk-1 was further supported by our observation that daf-2 clk-1 double mutants live much longer than either clk-1 or daf-2, and can live up to five times longer than the wild type (30). This result contrasts with the result of combining age-1 and daf-2, which, by genetic and molecular criteria, appear to function in the same pathway. In this case, the double mutants age-1(hx546); daf-2(e1370) do not live longer than daf-2(e1370) (28).
daf-16 Does Not Suppress the Long Life of eat-2.
To test whether CR extends life span by the same mechanism as the dauer genes, we tested whether daf-16 could suppress the long life of ad465, the reference allele of eat-2. The results of these experiments are very similar to those for daf-16(m26); clk-1(e2519) double mutants. daf-16(m26); eat-2(ad465) double mutants live slightly shorter than eat-2(ad465) (Fig. 2B). However, eat-2(ad465) mutants live 34% longer than the wild type and daf-16(m26); eat-2(ad465) double mutants live 36% longer than daf-16(m26).
eat-2; daf-2 Double Mutants Live Longer Than Either daf-2 or eat-2 Mutants.
The fact that daf-16 does not suppress the long life of eat-2 suggests that the dauer mutations and eat mutations lengthen life span by different mechanisms. One prediction of this observation is that, like daf-2(e1370) clk-1(e2519) double mutants, eat-2(ad465); daf-2(e1370) double mutants should live longer than animals carrying the individual mutations that compose these strains. This expectation is exactly what we find (Fig. 3A).
eat-2; clk-1 Double Mutants Live No Longer Than eat-2 Mutants.
To examine whether food restriction and clk-1 mutations lengthen life span by a similar mechanism, we examined the life span of eat-2(ad465); clk-1(e2519) double mutants. We find that eat-2; clk-1 double mutants do not live longer than either of the single mutants (Fig. 3B). This result is similar to the interaction of age-1 and daf-2, which function in the same pathway to determine life span. Thus, these results suggest that eat-2 and clk-1 mutations may regulate aging by affecting a common process.
DISCUSSION
eat Mutants Live Long Because of Food Restriction.
We find that most eat mutations significantly lengthen life span. Mutations in eat-2 can lengthen life span by more that 50%. In the cases of eat-2 and eat-6, life span appears to be correlated with the severity of the eating defect. These results are all consistent with the long life of eat mutants being caused by CR. Because eat mutations are believed to affect feeding behavior by disrupting the pharyngeal muscle or the nervous system, it was possible that eat mutants would live long because of pleiotropic effects on muscles and/or the nervous system. However, we find that mutations in 14 unc genes that do not affect pharyngeal pumping do not lengthen life span. Of all of the unc mutations examined, only mutations in the gene unc-26 lengthen life span. However unc-26 mutations lead to feeding defects and they probably have increased life spans because of CR. The results with unc mutants indicate that defects in body-wall muscles or the extrapharyngeal nervous system do not affect life span. They also indicate that decreased movement per se does not affect life span.
Many Genes Affect Life Span.
We have found that mutations in 7 of 10 genes that cause a feeding defect can lengthen life span. Because Avery (34) has estimated that there are at least 60 such genes that can mutate to give a feeding defect, mutations that extend life span by restricting food intake should be isolated at high frequency in genetic screens for long-lived mutants. We have also found that many unc strains harbor background mutations that extend life span. It is not clear whether these mutations affect feeding behavior; however, they do not affect developmental rates or cause dauer-formation defects (unpublished data).
Food Restriction and Dauer Mutations Lengthen Life Span by Distinct Mechanisms.
It has been hypothesized that CR may lengthen life span by affecting metabolism (reviewed in refs. 8 and 9). Because daf-2 encodes an insulin receptor-like molecule, and insulin regulates metabolism in mammals, it has been suggested that there may be parallels between life extension by CR and by daf-2 mutations (22). However, by two different criteria, the results of our aging studies strongly suggest that daf-2 mutations lengthen life span by a mechanism distinct from that of food restriction. This interpretation is supported by the phenotype of eat-2; daf-2 double mutants. eat-2 mutations appear to have no effect on the Daf-c phenotype of daf-2 mutants, consistent with these genes acting in different pathways. daf-2 mutants also have very different visible effects on metabolism than eat mutants. daf-2 mutants accumulate fat in the intestine, giving these mutants a very dark appearance (22). eat mutants, on the other hand, appear very pale, probably indicating reduced intestinal stores of fat. eat-2; daf-2 double mutants have an intermediate appearance: they are paler than daf-2 and darker than eat-2, suggesting that these two genes affect metabolism in different and perhaps antagonistic ways.
CR and clk-1 Mutations May Extend Life Span by a Similar Process.
Our results suggest that eat-2 and clk-1 mutations may affect life span by affecting a common process. eat-2 mutants and clk-1 mutants, however, have very different phenotypes. clk-1 mutations slow a range of timed phenomena, including the rate of embryonic and postembryonic development, as well as several behavioral rhythms (29). However, in spite of these wide-ranging defects, clk-1 mutants superficially have a wild-type appearance. We have interpreted the long life of clk-1 mutants to be the result of a slower rate of living, presumably concomitant with a reduced metabolic rate (20, 30). clk-1 encodes a small protein with an unknown, but evolutionarily conserved, biochemical function (31). Consistent with our interpretation of the function of clk-1, the yeast homologue of CLK-1 is localized to the inner membrane of mitochondria, where it is necessary for respiration (47, 48). On the other hand, eat-2 mutations have obvious deleterious effects. Mutations in the eat-2 gene are thought to affect primarily pharyngeal pumping rates, slowing pumping to 10–20% of normal (35). eat-2 mutants appear pale and small, two phenotypes that also are seen when wild-type worms are grown under low food concentrations (34). In spite of this, eat-2 mutants develop at an almost-wild-type rate. eat-2; clk-1 double mutants display the phenotypes of both mutations, developing at the same rate as clk-1 mutants and appearing pale and starved like eat-2 mutants, suggesting that only some aspects of their phenotype are relevant to the effect of these mutations on life span.
We hypothesize that mutations in both clk-1 and eat-2 genes may reduce energy production in the worm, and this is why they affect life span in a similar manner. eat-2 mutants are starved, which may lead, through some physiological feedback mechanism, to decreased metabolic rates. As the yeast homologue of CLK-1 is a mitochondrial protein, clk-1 mutations probably reduce metabolic rates more directly. However, if this is the case, clk-1 mutations also must lead to decreased energy requirements because unlike eat-2 mutants, clk-1 mutants are not starved.
Evidence is mounting that damage caused by reactive oxygen species (ROS) produced in the mitochondria may be a major cause of aging (8, 9). In other systems, CR has been shown to reduce oxidative stress produced by ROS (8, 9). If mutations in clk-1 and eat-2 genes reduce metabolic rates, this could lead to reduced rates of production of ROS, which in turn would lead to a slower accumulation of ROS-associated damage and slower aging.
The Relationship Between CR and Aging in Worms and Vertebrates.
We show that reducing food intake by eat mutations lengthens the life span of C. elegans. It is not well understood in C. elegans how reduced food intake lengthens life span, but by analogy to mammals it is likely that it is reduced caloric intake that lengthens life span. However, it has not been shown that this effect is caused by reduced caloric intake, as opposed to reduced intake of protein or some other nutrient. It also is not clear how similar the physiological response to reduced food intake is between worms and mammals. However, the fact that food restriction in worms, mammals, and many other types of organisms lengthens life span suggests that there may be important conserved physiological responses to reduced food intake across divergent phyla. The eat mutants provide excellent tools to help study these responses in a genetically tractable system.
Acknowledgments
We thank T. Barnes for lengthy discussions, C. Bénard and B. McCreight for carefully reading the manuscript, and D. Raizen, L. Avery, and J. Hodgkin for strains. Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health National Center for Research Resources. This work was supported by a grant to S.H. from the Medical Research Council of Canada and by a fellowship to B.L. from the Fonds pour la Formation de Chercheurs et l’Aide à la Recherche (FCAR), Québec.
ABBREVIATIONS
- CR
caloric restriction
- Eat
eating defective
- Unc
uncoordinated
- ROS
reactive oxygen species
References
- 1. McCay C M, Crowell M F, Maynard L A. J Nutr. 1935;10:63–79. [PubMed] [Google Scholar]
- 2.Weindruch R K, Walford R L. The Retardation of Aging and Disease by Dietary Restriction. Springfield, IL: Thomas; 1988. [Google Scholar]
- 3.Lane M A, Ball S S, Ingram D K, Cutler R G, Engel J, Read V, Roth G S. Am J Physiol. 1995;268:E941–E948. doi: 10.1152/ajpendo.1995.268.5.E941. [DOI] [PubMed] [Google Scholar]
- 4.Lane M A, Ingram D K, Roth G S. Age. 1997;20:45–56. doi: 10.1007/s11357-997-0004-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim M L, Aiken J M, Havighurst T, Hollander J, Ripple M O, Weindruch R. J Nutr. 1997;127:2293–2301. doi: 10.1093/jn/127.12.2293. [DOI] [PubMed] [Google Scholar]
- 6.Verdery R B, Ingram D K, Roth G S, Lane M A. Am J Physiol. 1997;273:E714–E719. doi: 10.1152/ajpendo.1997.273.4.E714. [DOI] [PubMed] [Google Scholar]
- 7.Weed J L, Lane M A, Roth G S, Speer D L, Ingram D K. Physiol Behav. 1997;62:97–103. doi: 10.1016/s0031-9384(97)00147-9. [DOI] [PubMed] [Google Scholar]
- 8.Sohal R S, Weindruch R. Science. 1996;273:59–63. doi: 10.1126/science.273.5271.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu B P. Free Radical Biol Med. 1996;21:651–668. doi: 10.1016/0891-5849(96)00162-1. [DOI] [PubMed] [Google Scholar]
- 10.Tuker V A. J Cell Physiol. 1965;65:393–403. doi: 10.1002/jcp.1030650313. [DOI] [PubMed] [Google Scholar]
- 11.Duffy P H, Feuers R, Nakamura K D, Leakey J, Hart R W. Chronobiol Int. 1990;7:113–124. doi: 10.3109/07420529009056963. [DOI] [PubMed] [Google Scholar]
- 12.Lane M A, Baer D J, Rumpler W V, Weindruch R, Ingram D K, Tilmont E M, Cutler R G, Roth G S. Proc Natl Acad Sci USA. 1996;93:4159–4164. doi: 10.1073/pnas.93.9.4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCarter R, Masoro E J, Yu B P. Am J Physiol. 1985;248:E488–E490. doi: 10.1152/ajpendo.1985.248.4.E488. [DOI] [PubMed] [Google Scholar]
- 14.Masoro E J, McCarter R J M. Aging (Milano) 1991;2:117–128. doi: 10.1007/BF03323988. [DOI] [PubMed] [Google Scholar]
- 15.Dulloo A G, Girardier L. Int J Obes Relat Metab Disord. 1993;17:115–123. [PubMed] [Google Scholar]
- 16.Ramsey J J, Roecker E B, Weindruch R, Kemnitz J W. Am J Physiol. 1997;272:E901–E907. doi: 10.1152/ajpendo.1997.272.5.E901. [DOI] [PubMed] [Google Scholar]
- 17.Rose M R. Evolutionary Biology of Aging. New York: Oxford Univ. Press; 1991. [Google Scholar]
- 18.Wood W B, Johnson T E. Curr Biol. 1994;4:151–153. doi: 10.1016/s0960-9822(94)00036-9. [DOI] [PubMed] [Google Scholar]
- 19.Jazwinski S M. Science. 1996;273:54–59. doi: 10.1126/science.273.5271.54. [DOI] [PubMed] [Google Scholar]
- 20.Hekimi S, Lakowski B, Barnes T M, Ewbank J J. Trends Genet. 1998;14:14–19. doi: 10.1016/S0168-9525(97)01299-7. [DOI] [PubMed] [Google Scholar]
- 21.Morris J Z, Tissenbaum H A, Ruvkun G. Nature (London) 1996;382:536–539. doi: 10.1038/382536a0. [DOI] [PubMed] [Google Scholar]
- 22.Kimura K D, Tissenbaum H A, Liu Y, Ruvkun G. Science. 1997;277:994–999. doi: 10.1126/science.277.5328.942. [DOI] [PubMed] [Google Scholar]
- 23.Lin K, Dorman J B, Rodan A, Kenyon C. Science. 1997;278:1319–1322. doi: 10.1126/science.278.5341.1319. [DOI] [PubMed] [Google Scholar]
- 24.Ogg S, Paradis S, Gottlieb S, Patterson G I, Lee L, Tissenbaum H A, Ruvkun G. Nature (London) 1997;389:994–999. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- 25.Gottlieb S, Ruvkun G. Genetics. 1994;137:107–120. doi: 10.1093/genetics/137.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. Nature (London) 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- 27.Larsen P L, Albert P S, Riddle D L. Genetics. 1995;139:1567–1583. doi: 10.1093/genetics/139.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dorman J B, Albinder B, Shroyer T, Kenyon C. Genetics. 1995;141:1399–1406. doi: 10.1093/genetics/141.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wong A, Boutis P, Hekimi S. Genetics. 1995;139:1247–1259. doi: 10.1093/genetics/139.3.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lakowski B, Hekimi S. Science. 1996;272:1010–1013. doi: 10.1126/science.272.5264.1010. [DOI] [PubMed] [Google Scholar]
- 31.Ewbank J J, Barnes T M, Lakowski B, Lussier M, Bussey H, Hekimi S. Science. 1997;275:980–983. doi: 10.1126/science.275.5302.980. [DOI] [PubMed] [Google Scholar]
- 32.Klass M R. Mech Ageing Dev. 1977;6:413–429. doi: 10.1016/0047-6374(77)90043-4. [DOI] [PubMed] [Google Scholar]
- 33.Hosono R, Nishimoto S, Kuno S. Exp Geront. 1989;24:251–264. doi: 10.1016/0531-5565(89)90016-8. [DOI] [PubMed] [Google Scholar]
- 34.Avery L. Genetics. 1993;133:897–917. doi: 10.1093/genetics/133.4.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raizen D M, Lee R Y N, Avery L. Genetics. 1995;141:1365–1382. doi: 10.1093/genetics/141.4.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Davis M W, Somerville D, Lee R Y N, Lockery S, Avery L, Fambrough D M. J Neurosci. 1995;15:8408–8418. doi: 10.1523/JNEUROSCI.15-12-08408.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Starich T A, Lee R Y N, Panzarella C, Avery L, Shaw J E. J Cell Biol. 1996;134:537–548. doi: 10.1083/jcb.134.2.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee R Y N, Lobel L, Hengartner M, Horvitz H R, Avery L. EMBO J. 1997;16:6066–6076. doi: 10.1093/emboj/16.20.6066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hekimi S, Boutis P, Lakowski B. Genetics. 1995;141:1351–1364. doi: 10.1093/genetics/141.4.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brenner S. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sultson J, Hodgkin J. In: The Nematode Caenorhabditis elegans. Wood W B, editor. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. pp. 587–606. [Google Scholar]
- 42.Weindruch R, Walford R L, Fligiel S, Guthrie D. J Nutr. 1986;116:641–654. doi: 10.1093/jn/116.4.641. [DOI] [PubMed] [Google Scholar]
- 43.Chalfie M, White J. In: The Nematode Caenorhabditis elegans. Wood W B, editor. Plainview, NY,: Cold Spring Harbor Lab. Press; 1988. pp. 337–391. [Google Scholar]
- 44.Johnson T E. In: Invertebrate Models in Aging Research. Mitchell D H, Johnson T E, editors. Boca Raton, FL: CRC; 1984. pp. 59–93. [Google Scholar]
- 45.Murakami S, Johnson T E. Genetics. 1996;143:1207–1218. doi: 10.1093/genetics/143.3.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Malone E A, Inoue T, Thomas J H. Genetics. 1996;143:1193–1205. doi: 10.1093/genetics/143.3.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marbois B N, Clarke C F. J Biol Chem. 1996;271:2995–3004. doi: 10.1074/jbc.271.6.2995. [DOI] [PubMed] [Google Scholar]
- 48.Jonassen T, Proft M, Randezgil F, Schultz J R, Marbois B N, Entian K-D, Clarke C F. J Biol Chem. 1998;273:3351–3357. doi: 10.1074/jbc.273.6.3351. [DOI] [PubMed] [Google Scholar]