Abstract
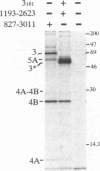
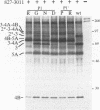
Cleavage at four sites (3/4A, 4A/4B, 4B/5A, and 5A/5B) in the hepatitis C virus polyprotein requires a viral serine protease activity residing in the N-terminal one-third of the NS3 protein. Sequence comparison of the residues flanking these cleavage sites reveals conserved features including an acidic residue (Asp or Glu) at the P6 position, a Cys or Thr residue at the P1 position, and a Ser or Ala residue at the P1' position. In this study, we used site-directed mutagenesis to assess the importance of these and other residues for NS3 protease-dependent cleavages. Substitutions at the P7 to P2' positions of the 4A/4B site had varied effects on cleavage efficiency. Only Arg at the P1 position or Pro at P1' substantially blocked processing at this site. Leu was tolerated at the P1 position, whereas five other substitutions allowed various degrees of cleavage. Substitutions with positively charged or other hydrophilic residues at the P7, P3, P2, and P2' positions did not reduce cleavage efficiency. Five substitutions examined at the P6 position allowed complete cleavage, demonstrating that an acidic residue at this position is not essential. Parallel results were obtained with substrates containing an active NS3 protease domain in cis or when the protease domain was supplied in trans. Selected substitutions blocking or inhibiting cleavage at the 4A/4B site were also examined at the 3/4A, 4B/5A, and 5A/5B sites. For a given substitution, a site-dependent gradient in the degree of inhibition was observed, with a 3/4A site being least sensitive to mutagenesis, followed by the 4A/4B, 4B/5A, and 5A/5B sites. In most cases, mutations abolishing cleavage at one site did not affect processing at the other serine protease-dependent sites. However, mutations at the 3/4A site which inhibited cleavage also interfered with processing at the 4B/5A site. Finally, during the course of these studies an additional NS3 protease-dependent cleavage site has been identified in the NS4B region.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bachmair A., Finley D., Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986 Oct 10;234(4773):179–186. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- Bartenschlager R., Ahlborn-Laake L., Mous J., Jacobsen H. Nonstructural protein 3 of the hepatitis C virus encodes a serine-type proteinase required for cleavage at the NS3/4 and NS4/5 junctions. J Virol. 1993 Jul;67(7):3835–3844. doi: 10.1128/jvi.67.7.3835-3844.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers T. J., Grakoui A., Rice C. M. Processing of the yellow fever virus nonstructural polyprotein: a catalytically active NS3 proteinase domain and NS2B are required for cleavages at dibasic sites. J Virol. 1991 Nov;65(11):6042–6050. doi: 10.1128/jvi.65.11.6042-6050.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckart M. R., Selby M., Masiarz F., Lee C., Berger K., Crawford K., Kuo C., Kuo G., Houghton M., Choo Q. L. The hepatitis C virus encodes a serine protease involved in processing of the putative nonstructural proteins from the viral polyprotein precursor. Biochem Biophys Res Commun. 1993 Apr 30;192(2):399–406. doi: 10.1006/bbrc.1993.1429. [DOI] [PubMed] [Google Scholar]
- Failla C., Tomei L., De Francesco R. Both NS3 and NS4A are required for proteolytic processing of hepatitis C virus nonstructural proteins. J Virol. 1994 Jun;68(6):3753–3760. doi: 10.1128/jvi.68.6.3753-3760.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuerst T. R., Niles E. G., Studier F. W., Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci U S A. 1986 Nov;83(21):8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushi S., Katayama K., Kurihara C., Ishiyama N., Hoshino F. B., Ando T., Oya A. Complete 5' noncoding region is necessary for the efficient internal initiation of hepatitis C virus RNA. Biochem Biophys Res Commun. 1994 Mar 15;199(2):425–432. doi: 10.1006/bbrc.1994.1246. [DOI] [PubMed] [Google Scholar]
- Grakoui A., McCourt D. W., Wychowski C., Feinstone S. M., Rice C. M. A second hepatitis C virus-encoded proteinase. Proc Natl Acad Sci U S A. 1993 Nov 15;90(22):10583–10587. doi: 10.1073/pnas.90.22.10583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grakoui A., McCourt D. W., Wychowski C., Feinstone S. M., Rice C. M. Characterization of the hepatitis C virus-encoded serine proteinase: determination of proteinase-dependent polyprotein cleavage sites. J Virol. 1993 May;67(5):2832–2843. doi: 10.1128/jvi.67.5.2832-2843.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grakoui A., Wychowski C., Lin C., Feinstone S. M., Rice C. M. Expression and identification of hepatitis C virus polyprotein cleavage products. J Virol. 1993 Mar;67(3):1385–1395. doi: 10.1128/jvi.67.3.1385-1395.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn C. S., Hahn Y. S., Braciale T. J., Rice C. M. Infectious Sindbis virus transient expression vectors for studying antigen processing and presentation. Proc Natl Acad Sci U S A. 1992 Apr 1;89(7):2679–2683. doi: 10.1073/pnas.89.7.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hijikata M., Kato N., Ootsuyama Y., Nakagawa M., Shimotohno K. Gene mapping of the putative structural region of the hepatitis C virus genome by in vitro processing analysis. Proc Natl Acad Sci U S A. 1991 Jul 1;88(13):5547–5551. doi: 10.1073/pnas.88.13.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hijikata M., Mizushima H., Akagi T., Mori S., Kakiuchi N., Kato N., Tanaka T., Kimura K., Shimotohno K. Two distinct proteinase activities required for the processing of a putative nonstructural precursor protein of hepatitis C virus. J Virol. 1993 Aug;67(8):4665–4675. doi: 10.1128/jvi.67.8.4665-4675.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houghton M., Weiner A., Han J., Kuo G., Choo Q. L. Molecular biology of the hepatitis C viruses: implications for diagnosis, development and control of viral disease. Hepatology. 1991 Aug;14(2):381–388. [PubMed] [Google Scholar]
- Hruby D. E., Guarino L. A., Kates J. R. Vaccinia virus replication. I. Requirement for the host-cell nucleus. J Virol. 1979 Feb;29(2):705–715. doi: 10.1128/jvi.29.2.705-715.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lin C., Chambers T. J., Rice C. M. Mutagenesis of conserved residues at the yellow fever virus 3/4A and 4B/5 dibasic cleavage sites: effects on cleavage efficiency and polyprotein processing. Virology. 1993 Feb;192(2):596–604. doi: 10.1006/viro.1993.1076. [DOI] [PubMed] [Google Scholar]
- Lin C., Lindenbach B. D., Prágai B. M., McCourt D. W., Rice C. M. Processing in the hepatitis C virus E2-NS2 region: identification of p7 and two distinct E2-specific products with different C termini. J Virol. 1994 Aug;68(8):5063–5073. doi: 10.1128/jvi.68.8.5063-5073.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manabe S., Fuke I., Tanishita O., Kaji C., Gomi Y., Yoshida S., Mori C., Takamizawa A., Yosida I., Okayama H. Production of nonstructural proteins of hepatitis C virus requires a putative viral protease encoded by NS3. Virology. 1994 Feb;198(2):636–644. doi: 10.1006/viro.1994.1075. [DOI] [PubMed] [Google Scholar]
- Mizushima H., Hijikata M., Tanji Y., Kimura K., Shimotohno K. Analysis of N-terminal processing of hepatitis C virus nonstructural protein 2. J Virol. 1994 Apr;68(4):2731–2734. doi: 10.1128/jvi.68.4.2731-2734.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizzi E., Tramontano A., Tomei L., La Monica N., Failla C., Sardana M., Wood T., De Francesco R. Molecular model of the specificity pocket of the hepatitis C virus protease: implications for substrate recognition. Proc Natl Acad Sci U S A. 1994 Feb 1;91(3):888–892. doi: 10.1073/pnas.91.3.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice C. M., Strauss J. H. Association of sindbis virion glycoproteins and their precursors. J Mol Biol. 1982 Jan 15;154(2):325–348. doi: 10.1016/0022-2836(82)90067-5. [DOI] [PubMed] [Google Scholar]
- Tomei L., Failla C., Santolini E., De Francesco R., La Monica N. NS3 is a serine protease required for processing of hepatitis C virus polyprotein. J Virol. 1993 Jul;67(7):4017–4026. doi: 10.1128/jvi.67.7.4017-4026.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukiyama-Kohara K., Iizuka N., Kohara M., Nomoto A. Internal ribosome entry site within hepatitis C virus RNA. J Virol. 1992 Mar;66(3):1476–1483. doi: 10.1128/jvi.66.3.1476-1483.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Sarnow P., Siddiqui A. Translation of human hepatitis C virus RNA in cultured cells is mediated by an internal ribosome-binding mechanism. J Virol. 1993 Jun;67(6):3338–3344. doi: 10.1128/jvi.67.6.3338-3344.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo B. J., Spaete R. R., Geballe A. P., Selby M., Houghton M., Han J. H. 5' end-dependent translation initiation of hepatitis C viral RNA and the presence of putative positive and negative translational control elements within the 5' untranslated region. Virology. 1992 Dec;191(2):889–899. doi: 10.1016/0042-6822(92)90264-p. [DOI] [PubMed] [Google Scholar]