Abstract
G protein signaling pathways regulate mitotic spindle positioning during cell division in many systems. In C. elegans embryos, Gα subunits act with the positive regulators GPR-1/2 and LIN-5 to generate cortical pulling forces for posterior spindle displacement during the first asymmetric division. GPR-1/2 are asymmetrically localized at the posterior cortex by PAR polarity cues at this time. Here we show that LIN-5 colocalizes with GPR-1/2 in one-cell embryos during spindle displacement. Significantly, we also find that LIN-5 and GPR-1/2 are localized to the opposite, anterior cortex in a polarity dependent manner during the nuclear centration and rotation movements that orient the forming spindle onto the polarity axis. The depletion of LIN-5 or GPR-1/2 results in decreased centration and rotation rates, indicating a role in force generation at this stage. The localization of LIN- 5 and GPR-1/2 is largely interdependent and requires Gα. Further, LIN-5 immunoprecipitates with Gα in vivo, and association is GPR-1/2 dependent. These results suggest that a complex of Gα /GPR- 1/2/LIN-5 is asymmetrically localized in response to polarity cues, and this may be the active signaling complex that transmits asymmetries to the force generation machinery during both nuclear rotation and spindle displacement.
Keywords: asymmetric division, nuclear rotation, spindle positioning, polarity, PAR-3, G protein signaling, LET-99, LIN-5, GPR-1/2
Introduction
Asymmetric divisions are essential for creating cell type diversity during development and for maintenance of stem cell lineages (Bellaiche and Gotta, 2005; Betschinger and Knoblich, 2004; Morrison and Kimble, 2006). During intrinsically asymmetric divisions, cell fate determinants become localized along a polarity axis. For proper partitioning of determinants to daughter cells, the mitotic spindle must align with the polarity axis, and the spindle is often displaced to result in daughters of unequal size. Alignment and displacement are mediated by astral microtubules, but the precise mechanisms by which these movements are coordinated with polarity cues are still being elucidated.
The conserved PAR polarity proteins regulate cell polarity and asymmetric division in many organisms. PAR proteins are asymmetrically localized during asymmetric divisions in embryonic cells of C. elegans and Drosophila and exhibit polarized distributions in many other cell types (reviewed in Bellaiche and Gotta, 2005; Betschinger and Knoblich, 2004; Macara, 2004). In addition, components of heterorimeric G protein signaling pathways influence spindle positioning in mammalian cells and are required for asymmetric division in Drosophila neuroblasts and C. elegans embryos (Bellaiche and Gotta, 2005; Betschinger and Knoblich, 2004; Macara, 2004). In these systems, G protein signaling is thought to be ligand and receptor-independent but requires several positive regulators including the GoLoco proteins Pins and Loco in Drosophila, LGN and AGS3 in mammals, and GPR-1/2 in C. elegans. All of these bind Gα subunits via their GoLoco domains and act as GDP dissociation inhibitors (Hampoelz and Knoblich, 2004).
In C. elegans embryos, PAR proteins establish cytoplasmic polarity and regulate spindle positioning during the first asymmetric division (reviewed in (Bellaiche and Gotta, 2005; Gönczy and Rose, 2005). During prophase, the pronuclear-centrosome complex moves toward the center (centration), and rotates 90° (nuclear rotation) to align the centrosomes on the anterior/posterior (A/P) axis defined by the PAR proteins. Posterior spindle displacement during metaphase and anaphase results in unequal cleavage to produce a larger anterior cell AB and smaller posterior cell, P1. In the P1 cell the nuclear-centrosome complex rotates to align with the PAR polarity axis. Biophysical studies indicate that these stereotypical nuclear-centrosome and spindle movements are driven by cortical pulling forces that act on astral microtubules. The forces switch from a net anterior force during centration/rotation to a net posterior force during metaphase, and both forces are regulated by PAR proteins (Grill et al., 2003; Labbe et al., 2004).
A heterotrimeric G protein signaling pathway acts downstream of the PARs to regulate posterior spindle displacement in the one-cell embryo, as well as nuclear rotation in the P1 cell. Two Gα proteins, GOA-1 and GPA-16, are partially redundant and are required for the majority of force generation during spindle displacement (Afshar et al., 2005; Colombo et al., 2003; Gotta and Ahringer, 2001; Gotta et al., 2003; Srinivasan et al., 2003; Tsou et al., 2003; Zwaal et al., 1996). RNA interference of GPR-1 and GPR-2 also results in a loss of force generation (hereafter referred to as GPR-1/2, as these are 96% identical). Gα subunits and GPR-1/2 are present in the cytoplasm, diffusely at centrosomes, and at the cortex. Gα cortical localization is uniform, but GPR-1/2 accumulate at higher levels at the posterior cortex beginning at metaphase (Colombo et al., 2003; Gotta and Ahringer, 2001;Gotta et al., 2003; Srinivasan et al., 2003; Tsou et al., 2003). GPR-1/2 asymmetry depends on the PAR proteins and this asymmetry is proposed to result in the posteriorly-directed pulling forces that mediate spindle displacement. The coiled-coil protein LIN-5 is also required for spindle displacement. LIN-5 localizes to centrosomes and the cortex, can associate with GPR-1/2, and is required for the cortical localization of GPR-1/2 (Gotta et al., 2003; Lorson et al., 2000; Srinivasan et al., 2003). LIN-5 shares weak homology to NuMA and Mud, which are microtubule binding proteins that associate with the Mammalian and Drosophila homologs of GPR-1/2, LGN and PINS respectively, to form a trimeric complex with Gα. It was thus proposed that LIN-5 may be a functional homolog of Mud and NuMA (Bowman et al., 2006; Du and Macara, 2004; Izumi et al., 2006; Siller et al., 2006). Mud localizes asymmetrically with the GPR-1/2 homolog PINS in Drosophila neuroblasts during division. However the precise relationship among LIN-5, GPR-1/2 and Gα in C. elegans has not been determined, and no asymmetry of LIN-5 in the one-cell embryo has been reported (Couwenbergs et al., 2004; Srinivasan et al., 2003).
The correct localization of GPR-1/2 also depends on the LET-99 protein. LET-99 is required for spindle positioning and is asymmetrically localized in a posterior cortical band pattern by the PAR proteins (Tsou et al., 2002; Tsou et al., 2003). Our previous studies showed that the highest levels of GPR-1/2 are posterior to the LET-99 band in wild type, and in let-99 mutants GPR-1/2 appeared more uniformly localized at the cortex. Interestingly, LET-99 also inhibits the overall cortical levels of GPR- 1/2 during prophase, and inhibition of Gα or GPR-1/2 suppressed the abnormal nuclear rocking movements exhibited by let-99 embryos during rotation (Tsou et al., 2003). These and other observations suggested that Gα signaling is involved in anteriorly-directed force generation during the time of centration and rotation. Consistent with this view, recent work shows that centration speed is reduced in Gα embryos (Goulding et al., 2007). However, whether Gá acts with GPR-1/2 and LIN-5 in centration is not known and no asymmetries in cortical GPR-1/2 localization have been reported at this stage (Couwenbergs et al., 2004; Srinivasan et al., 2003).
In this study we reexamine the localization of both LIN-5 and GPR-1/2 using quantitative analysis of staining intensities. We show for the first time that LIN-5 partially colocalizes with GPR- 1/2 at the cortex. Both LIN-5 and GPR-1/2 exhibit asymmetric cortical localization patterns that change during the cell cycle. These asymmetries correlate with force generation domains, and we show that both LIN-5 and GPR-1/2 are required for normal centration and rotation rates, in addition to their known role in posterior spindle displacement.
Results
LIN-5 and GPR-1/2 are enriched in the anterior during nuclear centration and rotation
To determine if any asymmetries of LIN-5 or GPR-1/2 localization are present during nuclear centration and rotation in one-cell embryos, we reexamined the localization of LIN-5 and GPR-1/2 in wild-type embryos using confocal microscopy of fixed specimens. Two different anti-LIN-5 antibodies (LIN-PAb, see Materials and Methods, and LIN-5 MAb (Srinivasan et al., 2003)) showed the same asymmetric staining patterns in wild-type embryos, which were absent in lin-5(RNAi) embryos (n>20;Fig. 1). Similarly all asymmetries revealed by anti-GPR staining were absent in gpr-1/2(RNAi) embryos (n>20; Fig. 1).
Figure 1.
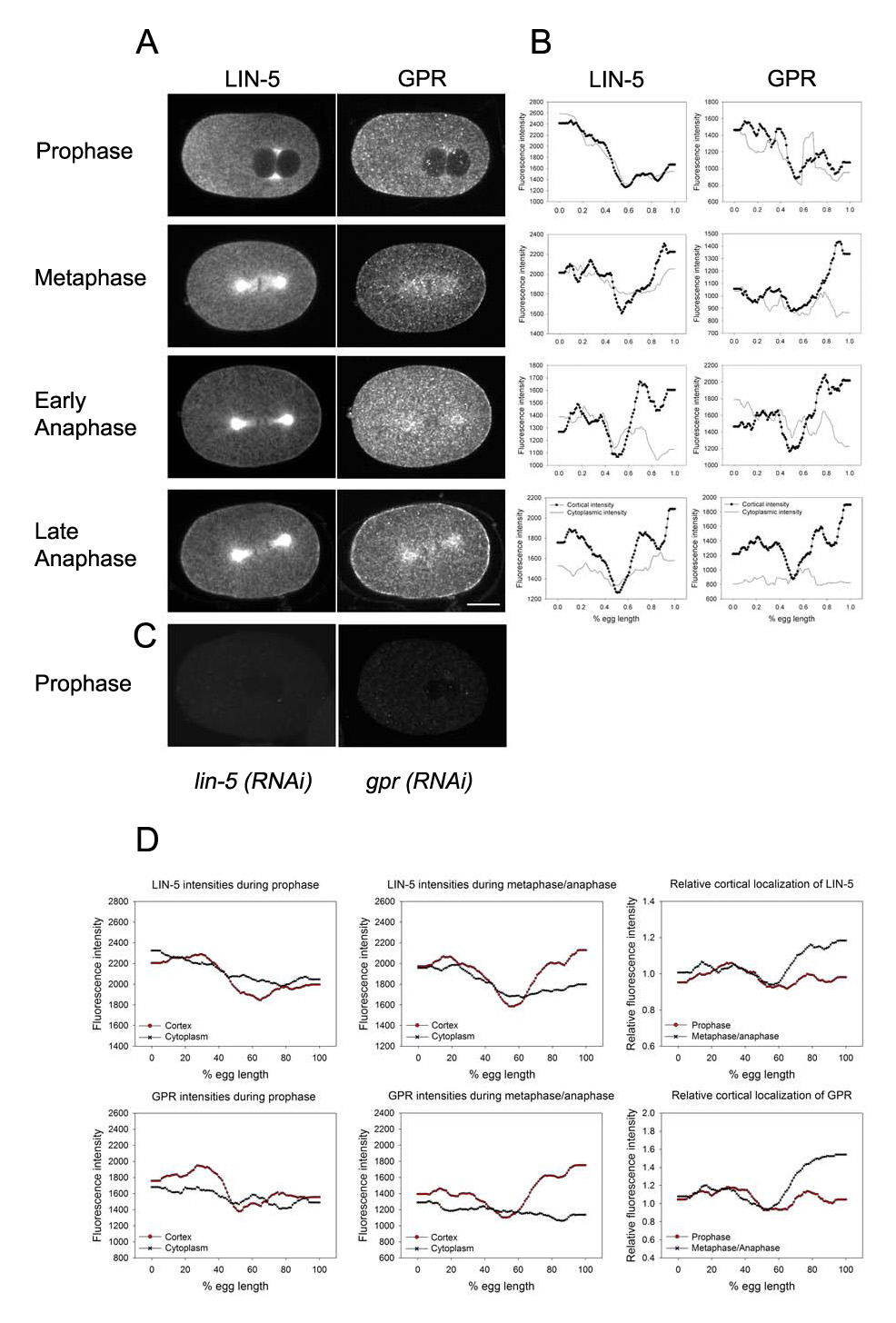
Asymmetric localization of LIN-5 and GPR-1/2. (A) Confocal images of wild-type one-cell embryos double-labeled with LIN-5 and GPR-1/2 antibodies at the stages indicated. Scale bars: 10µm. (B) Fluorescence intensity plots of the embryos shown in (A), from running averages of line scans. All embryos shown but early anaphase were stained with the LIN-5 PAb, which gives higher intensities. (C) Control staining for antibody specificity of LIN-5 PAb and GPR. (D) Average fluorescence intensity and relative cortical (cortical/cytoplasmic ratio) plots for wild-type embryos. n=10 cortices each, which differs from the text because only embryos stained with LIN-5 PAb were averaged.
During prophase of the first mitosis in centration and rotation stage embryos, LIN-5 did not appear enriched at the cortex relative to the underlying cytoplasm. However, the anterior cortex and cytoplasm stained more intensely for LIN-5 (Fig. 1A). To quantify this staining, we measured the fluorescence intensity of the upper and lower cortex of each embryo from anterior (0% egg length) to posterior (100% egg length); the intensity of the cytoplasm 1.5µm underneath the cortex was similarly measured. Representative traces are shown in Fig. 1B. Although both cortical and cytoplasmic LIN-5 intensities were somewhat discontinuous, most prophase embryos exhibited clear asymmetries: The highest levels of cortical LIN-5 staining were at the anterior from 0–30% egg length (33/38 cortices, n=19 embryos; note that in some embryos the upper and lower cortex showed different patterns and thus the number of cortices, rather than embryos, was scored). In these embryos, LIN-5 staining intensities began to decrease at about 40% egg length. LIN-5 cytoplasmic intensities showed similar anterior enrichment and the traces closely matched those of cortical levels in most cases (Fig. 1B, D; 30/38 traces). Averaged plots of cortical and cytoplasmic intensities for embryos stained with LIN-PAb are shown in Fig. 1D, and the differences between anterior and posterior intensities are statistically significant (Table 1). The averaged plot also shows a bipolar appearance rather than a simple anterior to posterior gradient: the region from ~50–70% egg length had the lowest average staining intensities. We note that although the average levels from 50–70% egg length are not statistically different than the posterior 80–100%, most individual cortices showed such a bipolar pattern (Fig. 1B; n = 25 bipolar cortices, 8 anterior gradient cortices). The lack of significance in the average intensities is potentially due to variation in the exact position of the lowest staining region, in addition to the inclusion of the few embryos with an anterior gradient staining pattern.
Table 1.
Comparison of staining intensities in wild-type and mutant embryos.
| Stage | Typea | Protein | Genotype | Range (% egg length) | Intensity (Mean±SD) | p valueb |
|---|---|---|---|---|---|---|
| Prophase | Cortex | LIN-5 | Wild type | 0–30% | 2244.09±141.61 | |
| 80–100% | 1977.17±239.12 | 0.0071* | ||||
| gpr (RNAi) | 0–30% | 1971.68±242.26 | 0.0053 | |||
| 80–100% | 1447.85±267.70 | 9.86E-05 | ||||
| let-99 (RNAi) | 0–30% | 2016.04±349.71 | 0.0638 | |||
| 80–100% | 1671.06±251.37 | 0.0058 | ||||
| GPR | Wild type | 0–30% | 1831.67±260.67 | |||
| 80–100% | 1562.28±279.97 | 0.039* | ||||
| let-99 (RNAi) | 0–30% | 2025.43±598.03 | 0.3568 | |||
| 80–100% | 1778.08±436.57 | 0.1985 | ||||
| Ratio | LIN-5 | Wild type | 0–100% | 0.9756±0.0294 | ||
| 80–100% | 0.9758±0.0597 | |||||
| gpr (RNAi) | 0–100% | 0.8887±0.0455 | 4.37E-05 | |||
| let-99 (RNAi) | 0–100% | 1.0127±0.0493 | 0.0438 | |||
| 80–100% | 1.0470±0.0595 | 0.0075 | ||||
| GPR | Wild type | 0–100% | 1.0705±0.0559 | |||
| let-99 (RNAi) | 0–100% | 1.3374±0.0938 | 2.37E-07 | |||
| Metaphase /Anaphase | Cortex | LIN-5 | Wild type | 0–30% | 2010.05±215.34 | |
| 80–100% | 2064.01±208.92 | 0.5766* | ||||
| gpr (RNAi) | 80–100% | 1647.12±182.04 | 0.0002 | |||
| GPR | Wild type | 0–30% | 1410.19±255.49 | |||
| 80–100% | 1677.28±236.63 | 0.0144* | ||||
| let-99 (RNAi) | 0–30% | 1909.60±584.56 | 0.0156 | |||
| 80–100% | 1815.08±588.19 | 0.4680 | ||||
| Ratio | LIN-5 | Wild type | 0–30% | 1.0334±0.1131 | ||
| 80–100% | 1.1840±0.0797 | 0.0029* | ||||
| gpr (RNAi) | 80–100% | 0.9571±0.0472 | 3.87E-07 |
Staining intensities were quantified at the cortex and cytoplasm as described in Methods. Ratio is the cortical intensity/cytoplasmic intensity.
p values were calculated between the wild type and the mutant for the same % egg length, except those marked with an asterick, which were compared to the wild type intensity for 0–30% egg length. Bolded p values are statistically significant (p<0.05)
In prophase embryos double labeled for LIN-5 and GPR-1/2, the cortical traces for the two proteins showed very similar patterns in individual embryos (9/10; Fig. 1B), and the average plot of GPR-1/2 cortical staining revealed a bipolar pattern with the highest levels at the anterior. Although cytoplasmic GPR-1/2 staining was higher in the anterior in some cases (5/10 traces), on average the pattern was more uniform than for LIN-5 cytoplasmic intensities (Fig. 1D). In addition, GPR-1/2 cortical intensities were typically higher than cytoplasmic intensities. This higher cortical to cytoplasmic intensity ratio for GPR-1/2 compared to LIN-5 (Fig. 1D, relative cortical signal) leads to the more obvious cortical appearance of GPR-1/2 detected by eye. It is not clear whether this cortical to cytoplasmic ratio has biological meaning (see below and Discussion). However, we found that the ratio consistently correlated with the qualitative assessment of the pattern as observed by eye, which was not always the same as the pattern detected by quantification of cortical staining intensities. We therefore present both cortical staining intensities and cortical to cytoplasmic ratios throughout this study, as it aids in comparisons between the images and the quantitative data, as well as in comparisons to previous qualitative analyses (Colombo et al., 2003; Gotta et al., 2003; Srinivasan et al., 2003).
We conclude that LIN-5 and GPR-1/2 colocalize at the cortex in an asymmetric fashion during centration and rotation. Significantly, this is the first report of asymmetric localization of positive regulators of G protein signaling at this stage, and could thus explain how forces are asymmetrically regulated during centration and rotation.
LIN-5 and GPR-1/2 are required for nuclear centration and rotation
The localization of LIN-5 and GPR-1/2 at the anterior cortex during prophase suggests that these proteins could play a role in cortical pulling forces at this time. One previous study reported incomplete rotation in some gpr-1/2(RNAi) embryos, but rotation defects in lin-5 embryos have not been reported (Colombo et al., 2003; Gotta et al., 2003; Srinivasan et al., 2003). Therefore, to determine if depletion of these proteins affects force generation, we quantitatively analyzed the rates of centration and rotation. Centration was measured in lin-5(ev571ts), gpr-1/2(RNAi) and wild-type embryos using DIC timelapse video-microscopy. The speed of centration was reduced significantly in gpr-1/2 and lin-5 embryos relative to wild type (Fig. 2A). Using DIC time-lapse, the trajectory of the centrosomes during rotation was difficult to follow, especially in the mutant backgrounds. Therefore to track rotation movements more accurately, RNA interference of GPR-1/2 was carried out in the background of the TH32 strain, which carries GFP::γ-tubulin to visualize the centrosomes and GFP::histone to visualize DNA. Two different centrosome phenotypes were observed over the time course of gpr-1/2(RNAi) from 30 to 49hrs: embryos in which the starting centrosome axis was perpendicular to the A/P axis as in wild type (8/15), and embryos in which the centrosomes were already aligned within 40° of the A/P axis at the time of pronuclear meeting (7/15). In this latter group, the male pronucleus was at a lateral instead of posterior position at the time of pronuclear meeting in several embryos, which could explain why the centrosomes were mispositioned. The proportion of embryos with this centrosome mispositioning phenotype increased with longer RNAi times (5/5 embryos filmed at 40–49 hr post treatment, versus 2/10 at 30–36hrs). In addition, 6/7 of these embryos exhibited completely symmetric anaphase spindle movements, while 8/8 of the embryos with normally positioned centrosomes at the time of pronuclear meeting later exhibited a slight posterior spindle displacement at metaphase/anaphase. Thus, the abnormal centrosome positioning phenotype appears to be the stronger loss of function phenotype.
Figure 2.
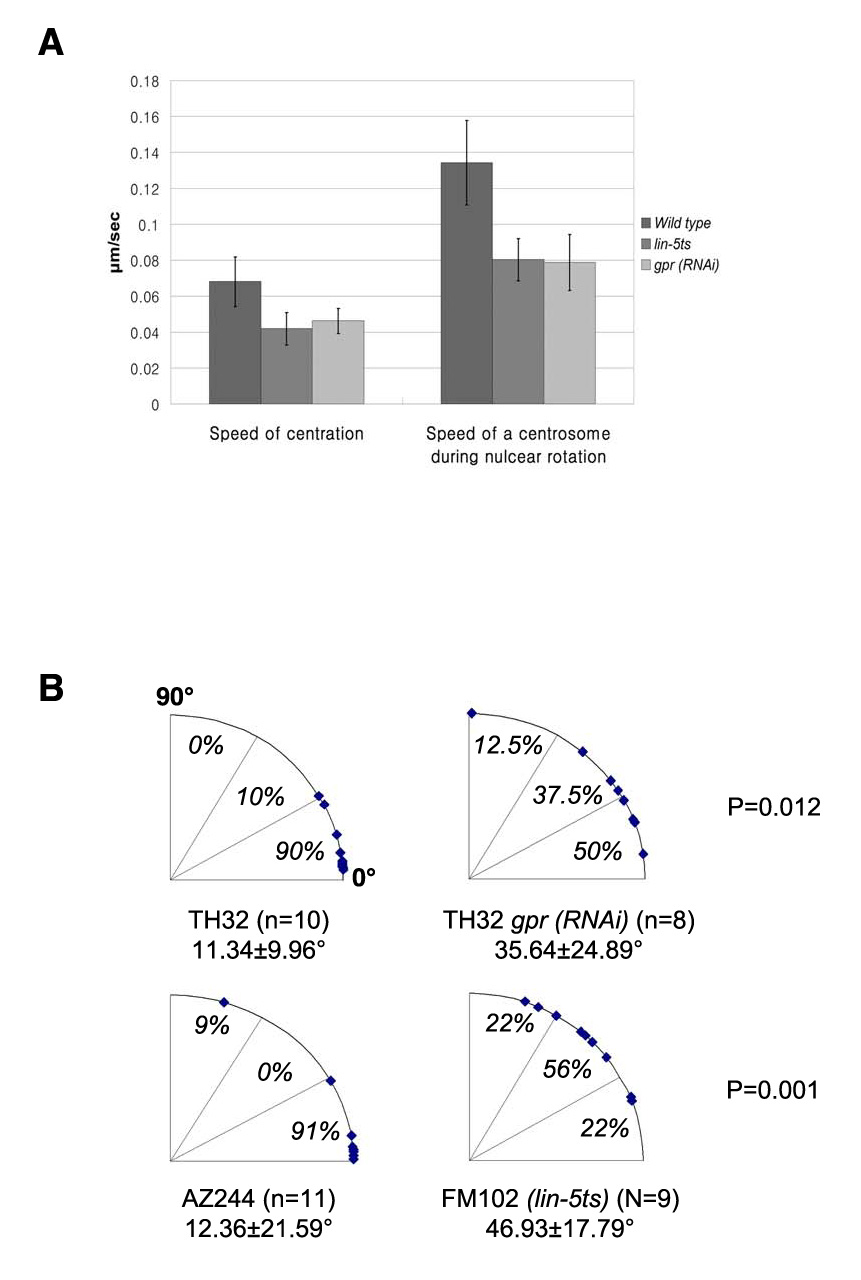
Defective centration and nuclear rotation in lin-5 and gpr-1/2 embryos. A) Centration and nuclear rotation are slower in lin-5 and gpr-1/2 embryos. Speeds of centration in wild type (0.068± 0.014µm/s, n=10), lin-5ts (ev571ts) (0.042± 0.009µm /s, n=9), and gpr-1/2 (RNAi) (0.046± 0.007µm /s, n=9) embryos. Speed of a centrosome during nuclear rotation in wild type (0.134± 0.024µm/s), FM102 (lin-5ts) (0.080± 0.012µm /s, n=9), and TH32 gpr-1/2 (RNAi) (0.079± 0.017µm /s, n=8,) embryos. Note that TH32 (n=10) and AZ244 (n=9) data were combined to calculate the speed of a centrosome in wild type because the results are not statistically different (p=0.138). Error bars indicate SD. P<0.002 for wild type versus lin-5 and gpr-1/2 in centration and P≪0.001 in nuclear rotation. B) Quantification of the extent of nuclear-centrosome rotation at NEB in lin-5 and gpr-1/2 embryos. Each dot represents a single embryo, with 0° indicating complete rotation of the complex onto the A/P axis. The average rotation angle ± SD and p values for wild type versus lin-5ts and gpr-1/2 (RNAi) are also given.
In the gpr-1/2(RNAi) embryos in which the centrosomes started in the normal wild-type position, the movements of the centrosomes were measured during nuclear rotation. Centrosome movements showed a normal rotational trajectory, but were slower on average than in wild type (Fig. 2A). In addition, in many embryos rotation was not complete rotation prior to NEB (Fig.2B). lin-5(RNAi) embryos showed a high proportion of embryos in which the centrosomes were already aligned (9/10) within 40° of the A/P axis; this combined with the presence of extra pronuclei resulting from the lin-5 meiotic defect (Lorson et al., 2000) made it difficult to score centrosome movements even in the TH32 background. Therefore, a lin-5 (ev571)ts; GFP:: α-tubulin strain was examined at non-permissive temperature. lin-5ts embryos showed a similar reduction in rotation rate as seen in gpr-1/2 embryos, as well as incomplete nuclear rotation (Fig. 2A, B). These results clearly demonstrate a role for LIN-5 and GPR-1/2 in normal centration and nuclear rotation in the one-cell embryo and support the model that the asymmetry of cortical LIN-5 and GPR-1/2 localization results in anteriorly-directed force generation at this stage. As noted above for gpr-1/2(RNAi), all of the lin-5 embryos in which rotation was examined still showed weakly asymmetric posterior spindle displacement, even though spindle oscillations were greatly reduced, suggesting that these embryos were not completely depleted of the protein. Thus, the partial rotation failure observed could represent either incomplete inactivation or the presence of another pathway that functions partially redundantly with LIN-5 and GPR-1/2. The mispositioning of the centrosomes prior to centration precluded distinguishing between these two possibilities.
Asymmetric localization of LIN-5 and GPR-1/2 from metaphase through division
GPR-1/2 were previously shown to be localized to the posterior cortex by metaphase (Colombo et al., 2003; Gotta et al., 2003; Tsou et al., 2003). To determine if LIN-5 colocalizes with GPR-1/2 at this stage also, and when the pattern changes from the anterior enrichment described above, we examined LIN-5 and GPR-1/2 localization throughout the rest of the cell cycle.
In prometaphase embryos, LIN-5 cortical and cytoplasmic staining patterns were similar to those at prophase (n= 5). At metaphase, posterior cortical enrichment of LIN-5 became apparent by visual inspection (Fig. 1A, metaphase). However, quantification showed that most cortices exhibited bipolar staining patterns. In most embryos, the posterior intensities were the same (11/28) or higher than anterior intensities (7/28 cortices; Fig. 1A, B), but some embryos exhibited bipolar patterns with higher anterior levels or anterior enrichment only (10/28), as in prophase stage embryos. Cytoplasmic LIN-5 was more enriched at the anterior in most metaphase stage embryos (20/28 cortices). Thus, the cortical to cytoplasmic ratio for LIN-5 was greater at the posterior for most embryos (23/28 cortices; Fig. 1B); this explains why embryos at this stage appear to have a posterior enrichment of cortical staining, even though the absolute levels of LIN-5 at the posterior are the highest in only 1/4 of the embryos.
All anaphase embryos showed posterior enrichment or bipolar enrichment of cortical LIN-5 (8 and 11 respectively; Fig. 1A). Cortical intensity traces for most embryos were actually bipolar (33/38 cortices), but as at metaphase, the cortical to cytoplasmic ratio better reflected the enrichment patterns observed by eye (Fig. 1D shows an average plot for the subset of metaphase and anaphase embryos stained with LIN-5 PAb). Bipolar and posterior enrichment of LIN-5 persisted through telophase and into the early two-cell interphase stage (24/26 embryos). Thus overall, LIN-5 cortical intensities show a bipolar asymmetric pattern throughout most of the cell cycle, with a tendency towards higher anterior staining intensities during prophase, followed by a shift towards higher posterior staining intensities at metaphase/anaphase.
In double-labeled embryos, LIN-5 and GPR-1/2 appeared to co-localize at the cortex at all stages from metaphase through the early 2-cell stage (n=>20 embryos; Fig. 1A) based on visual inspection. However, GPR-1/2 cortical intensity traces revealed that levels at the posterior were higher than at the anterior in many embryos (18/21 cortices). The average levels of GPR-1/2 at the posterior cortex relative to the anterior were significantly different, while LIN-5 anterior and posterior cortices were not different when averaged (Fig.1D and Table 1). Thus, LIN-5 and GPR-1/2 extensively colocalize at the posterior, but their patterns are not identical. These results are consistent with the model that LIN-5 and GPR-1/2 form a complex that is active for signaling to the force generation machinery during posterior spindle displacement, and further suggests that it is the colocalized population that is significant for spindle positioning.
Asymmetric patterns of LIN-5 and GPR-1/2 depend on polarity cues
It was previously shown that the asymmetric posterior enrichment of GPR-1/2 depends on the PAR-3 polarity protein and on LET-99, which is itself localized by the PAR pathway (Colombo et al., 2003; Gotta et al., 2003; Tsou et al., 2003). Similarly, we found that cortical LIN-5 staining at metaphase/anaphase was much more uniform in both par-3 and let-99 embryos compared to wild type (Fig. 3 shows a representative embryo, 4B shows average plots of intensities).
Figure 3.
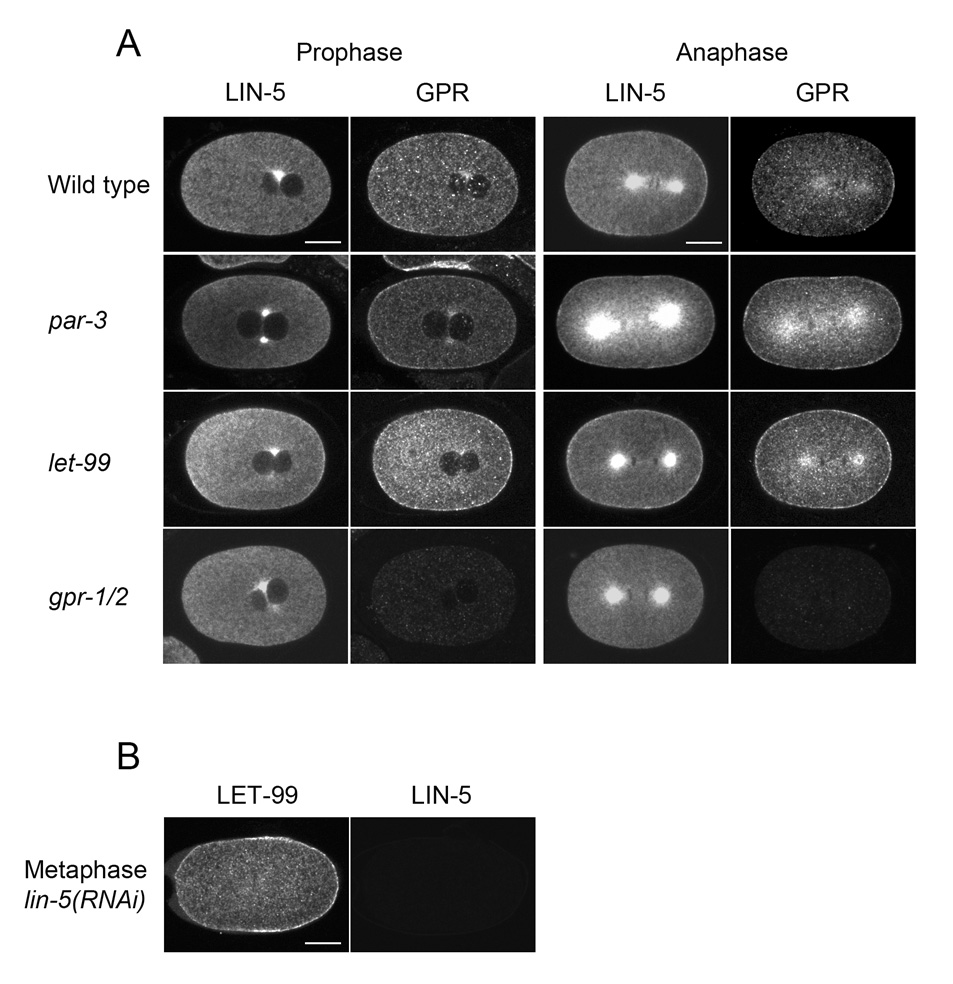
Cortical asymmetry of LIN-5 requires polarity and GPR-1/2. (A, B) Confocal images of embryos stained as indicated. Scale bars: 10µm.
To determine if the asymmetric anterior enrichment of GPR-1/2 or LIN-5 during centration/rotation is also polarity dependent, we examined staining in prophase par-3 embryos (Fig. 4). Cortical intensity traces of LIN-5 staining for individual par-3 embryos showed variable patchy patterns, which resulted in an average trace that was more uniform compared to wild type (Fig. 3A, middle). Cytoplasmic intensities were also uniform on average (Fig. 3A, middle). Thus, PAR-3 is required for the both cytoplasmic and cortical asymmetry of LIN-5 staining during prophase. Similarly, par-3 embryos stained for GPR-1/2 showed variable staining patterns. Although the averaged data shows a bias towards higher staining at the anterior, the bipolar pattern of the averaged intensities seen for wild type is not observed with par-3 (Fig. 3A, 4A left). Thus, PAR-3 is required for the normal pattern of GPR-1/2 at the cortex. However, these results suggest that other factors may also be involved in generating higher levels of GPR-1/2 at the anterior at this stage.
Figure 4.
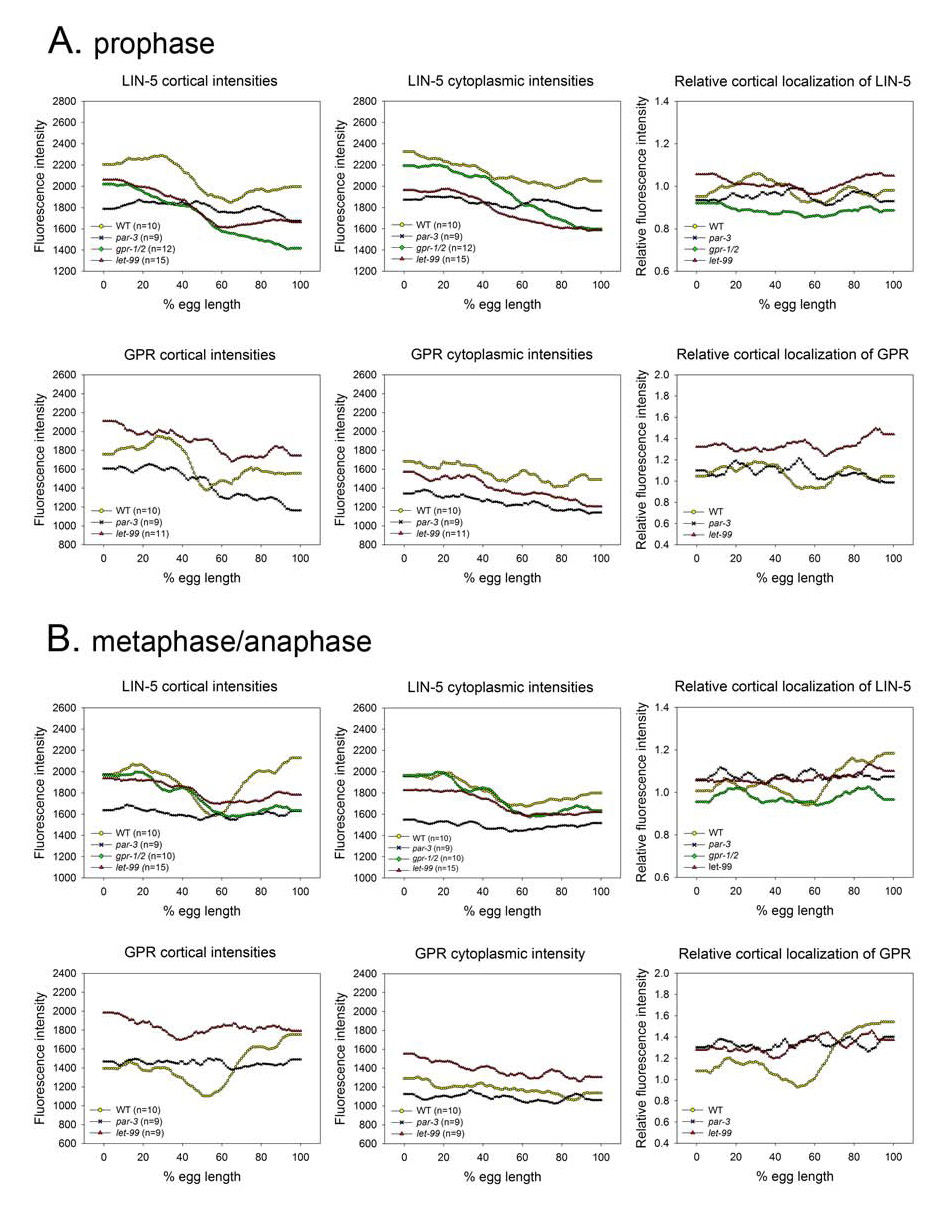
Average fluorescence intensity and relative cortical intensity plots for LIN-5 and GPR-1/2 in mutant embryos during prophase (A) and metaphase/anaphase (B). n= number of cortices examined. Wild-type plots are from Fig. 1.
Similarly, in let-99(RNAi) embryos during centration/rotation, cortical GPR-1/2 appeared more uniform, with a slight anterior to posterior gradient (Fig. 3, 4). In addition, the cortical to cytoplasmic ratio was significantly higher in let-99(RNAi) embyros than in wild type (Fig. 4), as previously reported for a let-99 mutant (Tsou et al., 2003). Cortical localization of LIN-5 was also more apparent by eye in let-99 embryos than in wild type. However, quantification of staining intensities showed that anterior enrichment was still present in let-99 embryos (Fig. 4). Surprisingly, the absolute levels of LIN-5 at the cortex were actually lower in let-99 embryos than in wild type. Thus, the higher “cortical” staining visible by eye once again appeared to reflect the cortical to cytoplasmic ratio, which was significantly higher in LIN-5 at the posterior. Thus, while absolute cortical levels of GPR-1/2 appear higher in let-99 embryos, absolute cortical levels of LIN-5 were not. Together these results confirm that LET-99 acts upstream of GPR-1/2 to inhibit their cortical localization; however, the observations that the absolute levels of LIN-5 are actually lower in let-99 embryos suggests that these proteins could also be affecting the trafficking of LIN-5 between the cortex and the cytoplasm or asters and that regulation is not simply at the level of cortical localization (see Discussion). In contrast, staining of lin-5 embryos for LET-99 showed that LET-99 is localized in a band pattern (Fig. 3), as in wild type and gpr-1/2(RNAi) embryos (Tsou et al., 2003).
Cortical LIN-5 asymmetry depends on GPR-1/2
It was previously shown that LIN-5’s cortical accumulation was reduced in gpr-1/2(RNAi) embryos after first division, but one-cell stage asymmetry was not examined (Srinivasan et al., 2003). Therefore we quantified LIN-5 staining intensities in gpr-1/2(RNAi) embryos. Cortical LIN-5 staining in prophase embryos was less apparent by eye, and average intensity plots confirmed that the cortical levels were reduced (Table 1). However, anterior cortical and cytoplasmic intensities were still asymmetric (Fig. 3, 4). At metaphase/anaphase, LIN-5 cortical staining was also greatly reduced but a weak cortical signal was detected in all embryos (n=23). However, the posterior enrichment of the cortical staining normally apparent by eye was abolished (20/23). Double-labeling confirmed that GPR-1/2 was completely depleted from the cortex (n= 23)(Srinivasan et al., 2003).
Based on the residual LIN-5 staining in these embryos and in previous studies (Srinivasan et al., 2003), we propose there are two pools of cortical LIN-5 detected in our studies: one that is GPR-1/2 dependent, and a second that is not. This model is also consistent with the different effects of loss of LET-99 activity on the localization of LIN-5 and GPR-1/2.
Evidence for a Gα/GPR-1/2/LIN-5 complex that regulates force generation
The cortical localization of GPR-1/2 depends on LIN-5, but also on Gα (Colombo et al., 2003; Gotta et al., 2003; Srinivasan et al., 2003; Tsou et al., 2003). To further explore the interdependencies of these proteins, we examined LIN-5 localization in Gα(RNAi) embryos at metaphase through the early two-cell stage, when cortical localization is most apparent.
The cortical localization of LIN-5 was greatly diminished but not absent in Gα(RNAi) embryos (n=17), and posterior enrichment of LIN-5 was abolished (16/17; Fig. 5A). However, cytoplasmic and centrosome staining of LIN-5 appeared wild type. As shown previously, GPR-1/2 were completely absent from the cortex in lin-5(RNAi) embryos (n= 8; Fig. 5B; Srinivasan et al., 2003). These results together with the analysis of gpr-1/2 and par-3 embryos suggest that LIN-5 forms a complex with Gα/GPR-1/2 at the cortex that responds to polarity cues.
Figure 5.
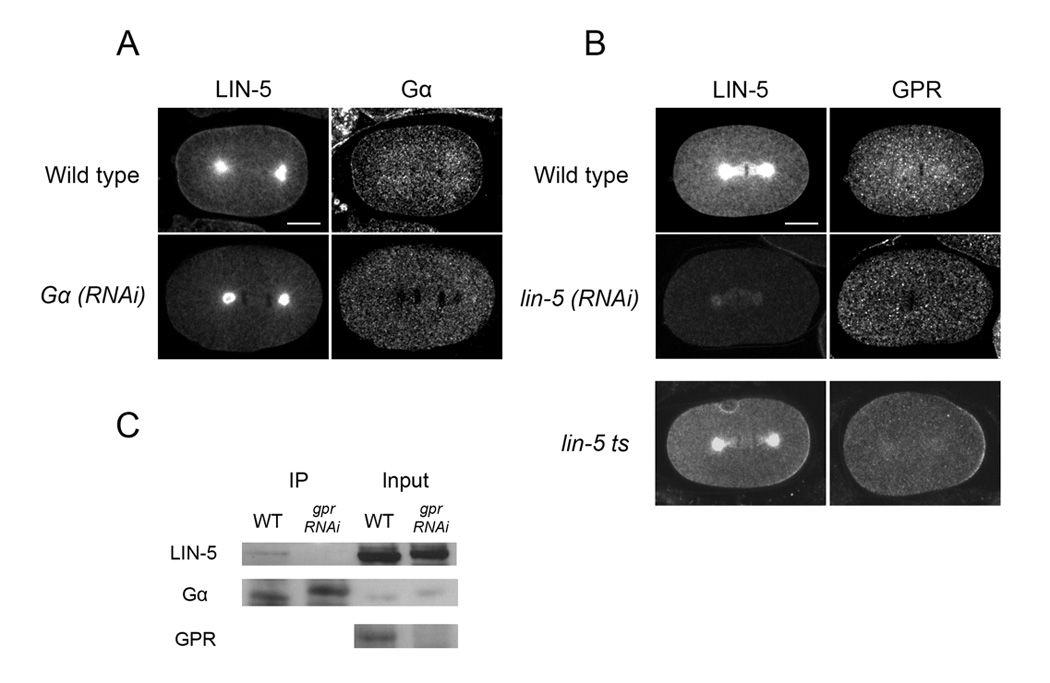
LIN-5 cortical localization depends on Gα. Confocal images of wild-type embryos double-labeled with the antibodies indicated. (A) Anaphase embryos. (B) Metaphase wild-type and lin- 5(RNAi) embryos and an anaphase lin-5(ev571ts) embryo. Scale bars: 10µm. (D) Western blots of Gα immunoprecipitation products. Anti- Gα antibody was used for immunoprecipitations (IP) from wild type and gpr-1/2(RNAi) embryo extracts. 1/40 dilution of input is shown for comparison. Products were probed with LIN-5 MAb, Gα and GPR antibodies.
Gα and LIN-5 associate with GPR-1/2 in vitro and in vivo (Colombo et al., 2003; Gotta et al., 2003; Srinivasan et al., 2003), but the presence of a complex containing all three had not been demonstrated at the start of our studies. To test for this, we carried out immunoprecipitation experiments using embryo extracts treated with the cross-linker DSP. LIN-5 co-immunoprecipitated with Gα in experiments using an antibody that recognizes both GOA-1 and GPA-16. LIN-5 was not present in control immunoprecipitations in which no primary antibody was used (not shown). Further, LIN-5 was not detected in immunoprecipitations of extracts made from gpr-1/2(RNAi) embryos (Fig. 5C). These results suggest that LIN-5 association with GOA-1/ GPA-16 is specific and that it depends on GPR-1/2, consistent with the formation of a complex.
In the course of analyzing LIN-5 and GPR-1/2 in the various mutant backgrounds, we found that GPR-1/2 and the mutant LIN-5 ts protein were both localized at the posterior cortex from metaphase on in lin-5(ev571ts) embryos, just as in wild type (n = 20, Fig. 5). Together these results show that the LIN-5(ev571ts) mutant protein is able to localize asymmetrically at the cortex in response to GPR-1/2 and polarity cues, and thus the defect in this mutant could be in transmitting this asymmetry to downstream effectors.
Discussion
In this report, we show that LIN-5 and GPR-1/2 exhibit asymmetric patterns of cortical localization that change during the cell cycle. While previous work revealed a critical role for LIN-5 in posterior spindle displacement, no asymmetry of LIN-5 localization was reported in the one-cell (Gotta et al., 2003; Lorson et al., 2000; Srinivasan et al., 2003). Our finding that LIN-5 localization is dynamic and asymmetry is often bipolar may explain the differences among the studies: We observed an overall anterior enrichment during centration/rotation, but quantification of cortical intensities revealed that many of these embryos exhibited the lowest staining intensities at 50–75% egg length, with higher staining intensities from 80–100% egg length. Similar, bipolar staining patterns were often observed during posterior spindle displacement, although at this time the posterior levels were highest in some cases. Quantification of cortical GPR-1/2 staining also revealed bipolar patterns in many embryos; however, during metaphase and anaphase GPR-1/2 was more enriched at the posterior relative to the anterior than was LIN-5. Thus, overall it is not the cortical staining pattern of LIN-5 per se, but the colocalized population of LIN-5 and GPR-1/2 that correlates with the direction of force generation during both rotation and posterior spindle displacement.
Previous studies showed that the GPR-1/2 and Gα signaling are required for the up-regulation of cortical forces during posterior spindle displacement (Colombo et al., 2003; Grill et al., 2003). The anterior enrichment of LIN-5 and GPR-1/2 during prophase could up-regulate cortical forces at this stage as well. Consistent with this hypothesis, we found that reduction in GPR-1/2 and LIN-5 result in reduced rates of centration, and slow and incomplete nuclear rotation. Interestingly, the rates of centration and rotation were similarly reduced in lin-5 and gpr-1/2 mutant embryos, even though LIN-5 still showed anterior enrichment in the cytoplasm in gpr-1/2 mutants. These observations support the model that it is the colocalized cortical population of LIN-5 and GPR-1/2, presumably a Gα/GPR- 1/2/LIN-5 complex, that is required for the generation of anteriorly-directed cortical forces. Gα mutants were likewise reported to have slower rotation in a recent report (Goulding et al., 2007); further, Gα mutants fail to rotate if extrinsic cell shape asymmetry is removed, as is seen with polarity mutants (Tsou et al., 2003). The failure to observe a complete lack of rotation in Gα, LIN-5 and GPR-1/2 depleted embryos that are normally shaped could be due to incomplete RNAi depletion. Alternatively, the role of Gα/GPR-1/2/LIN-5 in centration and rotation could be partially redundant with another mechanism. This second mechanism could involve the microtubule length-dependent pulling force mechanism proposed by (Kimura and Onami, 2005) which is proposed to result from force generators present on the cytoplasmic cytoskeleton rather than the cortex. We cannot distinguish between these two possibilities with current reagents, because we found that stronger depletion of LIN-5 and GPR-1/2 revealed an additional defect in which the centrosomes were already aligned with the A/P axis. Similarly, Gα RNAi embryos have centrosomes mispositioned on the A/P axis at the start of centration in more than half of the embryos ((Tsou et al., 2003)). In these mutants pronuclear meeting occurs at a position more anterior than in wild type (Couwenbergs et al., 2007; Park and Rose, unpublished), and we observed some embryos in which the male pronucleus was associated with the lateral rather than the posterior cortex. Thus, Gα signaling may also be required for male pronuclear attachment to the posterior cortex.
PAR polarity proteins and trimeric G protein signaling are conserved in regulating spindle movements in Drosophila, C. elegans and mammalian cells (Blumer et al., 2006; Bowman et al., 2006; Du and Macara, 2004; Fuja et al., 2004; Izumi et al., 2006; Siller et al., 2006). In Drosophila Gα forms a complex with the GPR-1/2 homolog PINS and the coiled-coil protein Mud, and all three are asymmetrically localized and regulate spindle positioning in neuroblasts. Similarly, in mammals, the binding of the coiled-coil protein NuMA to the GPR-1/2 homolog LGN potentiates the interaction of LGN with Gα and enhances cortical localization. Mud and NuMA share weak homology with each other and with LIN-5, and thus LIN-5 was proposed to be their functional counterpart in C. elegans(Bowman et al., 2006; Du and Macara, 2004; Izumi et al., 2006; Siller et al., 2006). In this report, we provide biochemical evidence for such a trimeric complex in C. elegans. Further, we show that the asymmetric and cortical localization of LIN-5 and GPR-1/2 is largely interdependent, consistent with the view that binding of LIN-5 to GPR-1/2 enhances its association with Gα, analogous to the findings for the mammalian complex. A concurrent study also reported the identification of a trimeric complex (Nguyen-Ngoc et al., 2007). In addition, that study and another recent report (Couwenbergs et al., 2007) provided evidence for association of LIN-5 and GPR-1/2 with subunits of the dynein motor complex, which have been implicated in providing motive force during spindle displacement (Pecreaux et al., 2006; Severson and Bowerman, 2003). Our study reveals a role for LIN-5 and GPR-1/2 in force generation during centration and nuclear rotation, which was previously shown to be dynein dependent as well (Gonczy et al., 1999; Schmidt et al., 2005). Thus, together these studies suggest that an asymmetric Gα/GPR-1/2/ LIN-5 cortical complex could be the active signaling module that directly regulates the force generation machinery during both nuclear rotation and spindle displacement. Significantly, NuMA can directly bind microtubules as well as the dynein/dynactin microtubule motor complex (Du and Macara, 2004; Du et al., 2002). We therefore propose that the Gα/GPR-1/2/LIN-5 complex could directly regulate microtubule behavior, or potentiate the cortical localization of LIN-5 to enable it to regulate microtubule-to-cortex interactions during nuclear and spindle positioning.
Although LIN-5 and GPR-1/2 appear to completely colocalize by visual inspection, our quantitative analysis revealed differences in the cortical and cytoplasmic staining patterns for these two proteins. For example, during metaphase LIN-5 and GPR-1/2 appeared completely colocalized at the posterior, and yet quantification revealed LIN-5 to have equally high cortical staining at both the anterior and posterior poles in many embryos. The pattern by eye more closely matched the cortical to cytoplasmic ratio, presumably because the eye can pick up small differences between the cortex and the underlying cytoplasm. It seems unlikely that this ratio itself can be read out by the cell, and thus we propose that there are two populations of LIN-5 at the cortex, a Gα/GPR-1/2-dependent pool and a GPR-1/2 independent pool of LIN-5. The observation that some asymmetry of LIN-5 staining at prophase remained in gpr-1/2(RNAi) embryos is consistent with this hypothesis, as is the result that LIN-5 cortical staining at later stages was greatly reduced but not abolished in gpr-1/2 and Gα embryos. This GPR-1/2 independent “cortical” LIN-5 population could be anchored at the cortex or be part of the cytoplasmic pool; because our cortical traces cover the outer 0.4um of the embryo, the intensities measured contain both the true cortex and the adjacent subcortical cytoplasm.
Our quantitative analysis of LIN-5 and GPR-1/2 staining also provides insight into the role of LET- 99 in regulating spindle positioning. We previously proposed that LET-99 acts downstream of the PAR pathway to inhibit the localization of GPR-1/2 and thus down-regulate G protein signaling at the cortex (Tsou et al., 2003). This was based on the increased cortical to cytoplasmic ratio of GPR-1/2 in let- 99(or81) mutant embryos. In this study, we confirm that the staining pattern of cortical GPR-1/2 is more uniform across the anterior-posterior axis in both let-99(RNAi) and par-3 embryos at prophase, consistent with our model. In addition, let-99(RNAi) had a greater effect on the prophase asymmetry of GPR-1/2 than of LIN-5, suggesting that LET-99 acts through GPR-1/2 to inhibit G protein signaling, rather through LIN-5. This differential effect is also consistent with the view that there are GPR-1/2 dependent and independent pools of LIN-5 detected in our cortical quantification. If two pools of LIN-5 exist, only the GPR-1/2 dependent LIN-5 would become uniformly localized in let-99 mutants, and the GPR-1/2 independent pool would remain anteriorly enriched. As in gpr-1/2 embryos, this anterior enrichment of LIN-5 in let-99 embryos does not appear to be sufficient for centration and rotation, presumably because it is no longer complexed with GPR-1/2. Interestingly, even though the highest levels of LET-99 are present in a band, more GPR-1/2 was seen uniformly around the entire cortex and no remaining asymmetry attributable to the PAR pathway was observed. These observations suggest that either the low levels of LET-99 present throughout the cortex are enough to inhibit GPR-1/2 accumulation, or that LET-99 in the cytoplasm could also play a role in GPR-1/2’s ability to accumulate at the cortex, or both. Indeed, we also observed that in let-99 embryos there was less staining intensity for LIN-5 at the cortex and in the subcortical cytoplasm, also pointing to a role for LET-99 in regulating the trafficking of LIN-5 to the cortex. Further experiments will be required to determine the detailed mechanism by which LET-99 regulates the localization of these proteins. Regardless, it is the apparent GPR-1/2 dependent pool of cortical LIN-5 whose asymmetry is dependent on LET-99 and which asymmetry correlates with the net forces acting on centrosomes in both wild-type and mutant embryos, supporting the model that a trimeric complex regulates force generation.
In summary, we present evidence that LIN-5 is part of a signaling complex that is conserved in spindle positioning in C. elegans, Drosophila and mammals (Bowman et al., 2006; Du and Macara, 2004; Izumi et al., 2006; Siller et al., 2006). In addition, we show for the first time that LIN-5 and GPR-1/2 are asymmetrically enriched at the anterior and function in centration and rotation, in addition to their known roles in spindle displacement. The dissection of the molecular mechanisms by which the dynamic patterns of these proteins are controlled by polarity cues and LET-99, and how LIN-5 and GPR-1/2 regulate dynein activity and force generation for spindle positioning, will be major areas of future research.
Materials and Methods
Strains and Culture
Worms were cultured on MYOB plates using standard methods. (Brenner, 1974; Church et al., 1995).Strains used were: N2, wild type Bristol variant; KK653, par-3(it71) unc-32(e189)/qC1; SV124, lin-5(ev571ts), TH32, unc-119(ed3) ddIs6[tbg-1::GFP + unc-119(+)]; ruIs32[unc-119(+) pie-1::GFP::H2B], AZ244, unc-119(ed3); ruIs57[pAZ147: pie-1/β-tubulin::GFP; unc-119(+)], FM102, lin-5(ev571ts) unc-119(ed3); ruIs57[pAZ147: pie-1/β-tubulin::GFP; unc-119(+)]. KK653 (Kemphues et al., 1988) was kindly provided by the Kemphues Lab, AZ244 and FM102 by the McNally Lab, TH32 by K. Oegema, and N2 and SV124 were obtained from the Caenorhabditis Genetics Center. SV124 was maintained at 16°C, and the others at 20°C.
Antibody generation and in situ immunolocalization
A 3’ fragment (1496-2466bp) of lin-5 cDNA (yk1155d09 from Y. Kohara, National Institute of Genetics, Japan) was cloned into pMAL-c2 (BioLabs) using PCR amplification. The MBP-LIN-5 fusion protein was expressed in bacteria, purified using Amylose resin (BioLabs), and injected into a rat (Covance). A full length goa-1 cDNA (amplified from the RB1 library) was cloned into pGEX-4T-1 (Amersham Bioscience). The GST-GOA-1 fusion protein was expressed in bacteria and purified using Glutathione Sepharose 4B resin (GE Healthcare) and injected into a rabbit (Covance). Antisera were purified using GST-LIN-5 or GST-GOA-1 fusion proteins cross-linked to Glutathione Sepharose 4B resin. The rat-polyclonal LIN-5 Ab generated is referred to as LIN-5 PAb, while the previously published mouse monoclonal Ab is LIN-5 MAb (Srinivasan et al., 2003). Rabbit-anti-LET-99 and rabbit-anti-GPR-2 antisera were previously generated (Tsou et al., 2003), but later bleeds for GPR-1/2 were purified and gave higher staining intensities in this study.
Immunolocalization was performed using standard freeze-fracture method followed by −20°C methanol fixation as described before (Tsou et al., 2002; DeBella et al., 2006). All primary antibodies were diluted 1:50 in PBST, and secondary antibodies (Jackson ImmunoResearch) were diluted 1:200 in PBST. DAPI staining was used to visualize DNA and to determine cell cycle stage. Specimens were mounted with Vectashield (Vector Laboratories) and observed using a 60X PLAPON NA 1.42 objective on an Olympus FV1000 Fluoview Laser Scanning Confocal Microscope. Z-stacks (0.2µm/section) were taken at the mid-focal plane of embryos using identical settings below saturation, using FV10-ASW software (ver. 1.5.0.14).
Quantification of staining intensities
Cortical and cytoplasmic fluorescence intensities were quantified using Image J software modules as described below (v. 1.37, NIH USA). A maximum intensity projection of stacks of 5 confocal images from a mid-focal plane was generated. A maximum filter (radius 1.5 pixels, 0.2µm) was applied to allow easier highlighting of the cortical versus subcortical region. Using Plot Profile, fluorescence intensities were obtained from a linescan drawn on the cortex, or in the cytoplasm 1.5µm below the cortex, from the middle of the anterior pole (0% egg length) to the middle of the posterior pole (100% egg length). Scatter plots of the raw data were smoothened using the running average program of SigmaPlot (v. 10.0, Systat Software; sampling proportion=0.1) to remove local fluctuation for analysis of patterns in individual embryos. For analysis of groups of embryos, the individual embryo lues along the A/P were averaged for each stage and genotype. Embryos were grouped by based on DAPIstaining of the nuclei and the position of nuclei and spindles into :prophase (from pronuclear meeting through rotation), prometaphase (nuclear envelope not visible), metaphase, and early and late anaphase (based on the extent of chromosome separation). Statistical tests of significance were made using the t test in SigmaPlot.
For presentation in figures, images were manually cut and aligned using Photoshop (Adobe). Contrast was adjusted to allow for better comparison of staining patterns carried out with antibodies with different staining intensities. However, for comparisons of wild type to RNAi depletions for staining controls, the images were not adjusted.
RNA interference
Sense and antisense RNAs were transcribed in vitro from full-length cDNA templates obtained by PCR amplification and annealed as described (Fire et al., 1998). L4 larvae were soaked in approximately 1.0mg/ml dsRNA solution for 18–24 hours at 20°C; for Gα depletion, a 1:1 mixture of goa-1 and gpa-16 dsRNA was used. After 30 hours recovery at 20°C, the progeny of soaked worms were analyzed. Bacterial RNAi feeding strains for let-99 (MRC geneservice ID: IV-6P07), lin-5 (II-5J10), and gpr-2 (III-5C03) were also used. L4 larvae were placed on bacteria and the embryos examined after 24–28 hours for let-99 RNAi and 30–34 hours for lin-5 RNAi at 20°C. For mass culture of gpr(RNAi) worms, synchronized L1 larvae were placed on bacteria, and the embryos obtained after 60 hours at 20°C. Phenotypes of all RNAi embryos were monitored using time-lapse video microscopy before use to ensure the effectiveness of the treatment, and by antibody staining for immunolocalization experiments.
Immunoprecipitations
Embryonic extracts were prepared as in (Afshar et al., 2005) except that cross-linking was carried out on embryos before lysis. For cross-linking, DSP (Pierce) was added to a final concentration of 1mM and the reaction carried out according to the manufacturer’s instructions. For immunoprecipitation assay, at least 400 µg of embryonic extract was incubated with 4 µl of anti-Gα antibody for two hours at 4°C with gentle rocking, followed by 30 µl of Protein A Sepharose CL-4B beads (GE healthcare) overnight at 4°C. The protein-bead complex was washed three times with 500µl of the ice cold reaction buffer. Associated proteins were eluted with SDS loading buffer, and analyzed by SDS PAGE and western blotting using standard protocols. GPR-1/2 and Gá primary antibodies were used at 1:2000 in PBST with 5% dry milk, and LIN-5 mAb at 1:5000. HRP conjugated secondary antibodies (1:5000 dilution) and ECL Plus (GE Healthcare) were used for protein detection.
Time-lapse videomicroscopy
To examine centration and rotation, one-cell embryos were mounted on slides without flattening (Rose and Kemphues, 1998) and recorded using DIC time-lapse video microscopy at room temperature of 23–25°C. To calculate the speed of centration, the migration distance of the center of the nuclear-centrosome complex from the time of pronuclear meeting to the time when nuclear rotation begins was measured.
For quantitative analysis of centrosome movements during nuclear rotation, live imaging of embryos expressing GFP:: α-tubulin or GFP::histone and GFP::γtubulin was performed using an Olympus BX60 epifluorescence microscope (UPlanF1 100x/1.30NA objective) and OpenLab software (ver. 3.1.7, Improvision). Images were taken every 5 seconds from pronuclear meeting to the end of the first cell cycle and then analyzed using ImageJ. The orientation of centrosomes at nuclear envelope breakdown (NEB) was determined by the angle of the centrosomal axis with respect to the anterior-posterior axis. The trajectory of a centrosome during 10 frames (50 sec.) before NEB was measured to calculate speed of a centrosome during nuclear rotation.
Acknowledgements
We thank S. van den Heuvel for antibodies, K. Kemphues and K. McNally for strains, Y. Kohara for cDNAs and J. Wu for help with constructs. Other strains were provided by the Caenorhabditis Genetics Center (funded by the NIH National Center for Research Resources). We are grateful to Dan Starr and Jui-ching Wu for comments on the manuscript, and the Rose and McNally labs for helpful discussions. This research was supported by NIH R01GM68744.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Afshar K, Willard FS, Colombo K, Siderovski DP, Gonczy P. Cortical localization of the Galpha protein GPA-16 requires RIC-8 function during C. elegans asymmetric cell division. Development. 2005;132:4449–4459. doi: 10.1242/dev.02039. [DOI] [PubMed] [Google Scholar]
- Bellaiche Y, Gotta M. Heterotrimeric G proteins and regulation of size asymmetry during cell division. Curr Opin Cell Biol. 2005;17:658–663. doi: 10.1016/j.ceb.2005.10.002. [DOI] [PubMed] [Google Scholar]
- Betschinger J, Knoblich JA. Dare to be different: asymmetric cell division in Drosophila, C. elegans and vertebrates. Curr Biol. 2004;14:R674–R685. doi: 10.1016/j.cub.2004.08.017. [DOI] [PubMed] [Google Scholar]
- Blumer JB, Kuriyama R, Gettys TW, Lanier SM. The G-protein regulatory (GPR) motif-containing Leu-Gly-Asn-enriched protein (LGN) and Gialpha3 influence cortical positioning of the mitotic spindle poles at metaphase in symmetrically dividing mammalian cells. Eur J Cell Biol. 2006;85:1233–1240. doi: 10.1016/j.ejcb.2006.08.002. [DOI] [PubMed] [Google Scholar]
- Bowman SK, Neumuller RA, Novatchkova M, Du Q, Knoblich JA. The Drosophila NuMA Homolog Mud regulates spindle orientation in asymmetric cell division. Dev Cell. 2006;10:731–742. doi: 10.1016/j.devcel.2006.05.005. [DOI] [PubMed] [Google Scholar]
- Colombo K, Grill SW, Kimple RJ, Willard FS, Siderovski DP, Gonczy P. Translation of polarity cues into asymmetric spindle positioning in Caenorhabditis elegans embryos. Science. 2003;300:1957–1961. doi: 10.1126/science.1084146. [DOI] [PubMed] [Google Scholar]
- Couwenbergs C, Labbe JC, Goulding M, Marty T, Bowerman B, Gotta M. Heterotrimeric G protein signaling functions with dynein to promote spindle positioning in C. elegans. J Cell Biol. 2007;179:15–22. doi: 10.1083/jcb.200707085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couwenbergs C, Spilker AC, Gotta M. Control of embryonic spindle positioning and Galpha activity by C. elegans RIC-8. Curr Biol. 2004;14:1871–1876. doi: 10.1016/j.cub.2004.09.059. [DOI] [PubMed] [Google Scholar]
- Du Q, Macara IG. Mammalian Pins is a conformational switch that links NuMA to heterotrimeric G proteins. Cell. 2004;119:503–516. doi: 10.1016/j.cell.2004.10.028. [DOI] [PubMed] [Google Scholar]
- Du Q, Taylor L, Compton DA, Macara IG. LGN blocks the ability of NuMA to bind and stabilize microtubules. A mechanism for mitotic spindle assembly regulation. Curr Biol. 2002;12:1928–1933. doi: 10.1016/s0960-9822(02)01298-8. [DOI] [PubMed] [Google Scholar]
- Fuja TJ, Schwartz PH, Darcy D, Bryant PJ. Asymmetric localization of LGN but not AGS3, two homologs of Drosophila pins, in dividing human neural progenitor cells. J Neurosci Res. 2004;75:782–793. doi: 10.1002/jnr.10874. [DOI] [PubMed] [Google Scholar]
- Gonczy P, Pichler S, Kirkham M, Hyman AA. Cytoplasmic dynein is required for distinct aspects of MTOC positioning, including centrosome separation, in the one cell stage Caenorhabditis elegans embryo. J Cell Biol. 1999;147:135–150. doi: 10.1083/jcb.147.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gönczy P, Rose LS. Asymmetric cell division and axis formation in the embryo. In: T. C. e. R. Community, editor. Wormbook. 2005. http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotta M, Ahringer J. Distinct roles for Galpha and Gbetagamma in regulating spindle position and orientation in Caenorhabditis elegans embryos. Nat Cell Biol. 2001;3:297–300. doi: 10.1038/35060092. [DOI] [PubMed] [Google Scholar]
- Gotta M, Dong Y, Peterson YK, Lanier SM, Ahringer J. Asymmetrically distributed C. elegans homologs of AGS3/PINS control spindle position in the early embryo. Curr Biol. 2003;13:1029–1037. doi: 10.1016/s0960-9822(03)00371-3. [DOI] [PubMed] [Google Scholar]
- Goulding MB, Canman JC, Senning EN, Marcus AH, Bowerman B. Control of nuclear centration in the C. elegans zygote by receptor-independent Galpha signaling and myosin II. J Cell Biol. 2007;178:1177–1191. doi: 10.1083/jcb.200703159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grill SW, Howard J, Schaffer E, Stelzer EH, Hyman AA. The distribution of active force generators controls mitotic spindle position. Science. 2003;301:518–521. doi: 10.1126/science.1086560. [DOI] [PubMed] [Google Scholar]
- Hampoelz B, Knoblich JA. Heterotrimeric G proteins: new tricks for an old dog. Cell. 2004;119:453–456. doi: 10.1016/j.cell.2004.10.025. [DOI] [PubMed] [Google Scholar]
- Izumi Y, Ohta N, Hisata K, Raabe T, Matsuzaki F. Drosophila Pins-binding protein Mud regulates spindle-polarity coupling and centrosome organization. Nat Cell Biol. 2006 doi: 10.1038/ncb1409. [DOI] [PubMed] [Google Scholar]
- Kemphues KJ, Priess JR, Morton DG, Cheng NS. Identification of genes required for cytoplasmic localization in early C. elegans embryos. Cell. 1988;52:311–320. doi: 10.1016/s0092-8674(88)80024-2. [DOI] [PubMed] [Google Scholar]
- Kimura A, Onami S. Computer simulations and image processing reveal length-dependent pulling force as the primary mechanism for C. elegans male pronuclear migration. Dev Cell. 2005;8:765–775. doi: 10.1016/j.devcel.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Labbe JC, McCarthy EK, Goldstein B. The forces that position a mitotic spindle asymmetrically are tethered until after the time of spindle assembly. J Cell Biol. 2004;167:245–256. doi: 10.1083/jcb.200406008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorson MA, Horvitz HR, van den Heuvel S. LIN-5 is a novel component of the spindle apparatus required for chromosome segregation and cleavage plane specification in Caenorhabditis elegans. J Cell Biol. 2000;148:73–86. doi: 10.1083/jcb.148.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macara IG. Par proteins: partners in polarization. Curr Biol. 2004;14:R160–R162. [PubMed] [Google Scholar]
- Morrison SJ, Kimble J. Asymmetric and symmetric stem-cell divisions in development and cancer. Nature. 2006;441:1068–1074. doi: 10.1038/nature04956. [DOI] [PubMed] [Google Scholar]
- Nguyen-Ngoc T, Afshar K, Gonczy P. Coupling of cortical dynein and Galpha proteins mediates spindle positioning in Caenorhabditis elegans. Nat Cell Biol. 2007 doi: 10.1038/ncb1649. [DOI] [PubMed] [Google Scholar]
- Pecreaux J, Roper JC, Kruse K, Julicher F, Hyman AA, Grill SW, Howard J. Spindle oscillations during asymmetric cell division require a threshold number of active cortical force generators. Curr Biol. 2006;16:2111–2122. doi: 10.1016/j.cub.2006.09.030. [DOI] [PubMed] [Google Scholar]
- Schmidt DJ, Rose DJ, Saxton WM, Strome S. Functional analysis of cytoplasmic dynein heavy chain in Caenorhabditis elegans with fast-acting temperature-sensitive mutations. Mol Biol Cell. 2005;16:1200–1212. doi: 10.1091/mbc.E04-06-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severson AF, Bowerman B. Myosin and the PAR proteins polarize microfilament-dependent forces that shape and position mitotic spindles in Caenorhabditis elegans. J Cell Biol. 2003;161:21–26. doi: 10.1083/jcb.200210171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siller KH, Cabernard C, Doe CQ. The NuMA-related Mud protein binds Pins and regulates spindle orientation in Drosophila neuroblasts. Nat Cell Biol. 2006 doi: 10.1038/ncb1412. [DOI] [PubMed] [Google Scholar]
- Srinivasan DG, Fisk RM, Xu H, van den Heuvel S. A complex of LIN-5 and GPR proteins regulates G protein signaling and spindle function in C elegans. Genes Dev. 2003;17:1225–1239. doi: 10.1101/gad.1081203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsou MF, Hayashi A, DeBella LR, McGrath G, Rose LS. LET-99 determines spindle position and is asymmetrically enriched in response to PAR polarity cues in C. elegans embryos. Development. 2002;129:4469–4481. doi: 10.1242/dev.129.19.4469. [DOI] [PubMed] [Google Scholar]
- Tsou MF, Hayashi A, Rose LS. LET-99 opposes Galpha/GPR signaling to generate asymmetry for spindle positioning in response to PAR and MES-1/SRC-1 signaling. Development. 2003;130:5717–5730. doi: 10.1242/dev.00790. [DOI] [PubMed] [Google Scholar]
- Zwaal RR, Ahringer J, van Luenen HG, Rushforth A, Anderson P, Plasterk RH. G proteins are required for spatial orientation of early cell cleavages in C. elegans embryos. Cell. 1996;86:619–629. doi: 10.1016/s0092-8674(00)80135-x. [DOI] [PubMed] [Google Scholar]


