Abstract
Werner syndrome (WS) is an autosomal recessive disorder characterized by genomic instability and the premature onset of a number of age-related diseases. The gene responsible for WS encodes a member of the RecQ-like subfamily of DNA helicases. Here we show that its murine homologue maps to murine chromosome 8 in a region syntenic with the human WRN gene. We have deleted a segment of this gene and created Wrn-deficient embryonic stem (ES) cells and WS mice. While displaying reduced embryonic survival, live-born WS mice otherwise appear normal during their first year of life. Nonetheless, although several DNA repair systems are apparently intact in homozygous WS ES cells, such cells display a higher mutation rate and are significantly more sensitive to topoisomerase inhibitors (especially camptothecin) than are wild-type ES cells. Furthermore, mouse embryo fibroblasts derived from homozygous WS embryos show premature loss of proliferative capacity. At the molecular level, wild-type, but not mutant, WS protein copurifies through a series of centrifugation and chromatography steps with a multiprotein DNA replication complex.
Werner syndrome (WS) is a rare autosomal recessive disorder characterized by the premature onset of a number of processes associated with aging (1, 2). The average age at death of WS patients is between 45 and 50 years, with the major causes of death being—as in normal individuals—myocardial infarction or malignancy. WS patients also manifest nonage-associated symptoms such as short stature, lack of the postadolescent growth spurt, and hypogonadism (1). In addition, the phenotype of cultured fibroblasts explanted from patients suffering from WS also suggests a parallel between WS and aging. The proliferative life-span of WS fibroblasts is reduced as compared with age-matched controls. Thus, WS cells behave like fibroblasts established from elderly individuals (3–5). WS cells also exhibit variegated chromosomal translocations and deletions, abnormalities that have been found in cultured WS skin fibroblasts as well as in lymphoblastoid cells (6, 7).
The gene responsible for WS (WRN) is located on the short arm of human chromosome 8 (8p11.1–21.1) (8–11) and was identified by positional cloning (12). The protein sequence predicted from this gene shows significant similarity to the DNA helicases (see below) and several mutations have been characterized, including mutations in splice acceptor sites, nonsense mutations, and genomic deletions of several exons encoding portions of the helicase domain (12, 13).
Significantly, the WRN gene product contains seven helicase consensus domains that are 34–38% identical to the Escherichia coli RecQ gene (14) and to the putative yeast helicase Sgs1p (15, 16). The Sgs1p yeast helicase is known to interact with type I and II topoisomerases and is required for genome stability in Saccharomyces cerevisiae (15–17). This interaction in yeast may represent an important clue as to the action of the WS helicase in mammalian cells. Type I topoisomerase, which nicks DNA creating a single-strand break, is involved in some aspects of replication and transcription. It has been identified as a cofactor of activator-dependent transcription by RNA polymerase II (18). In the fly, topoisomerase I is enriched in actively transcribed regions of the genome including the nucleolus, where transcription of rRNA by RNA polymerase I occurs (19). Interestingly, Sgs1p mutant yeast cells display a premature fragmentation of their nucleoli, suggesting an important function of this RecQ helicase family member in the nucleolus (20). Mammalian type II topoisomerase, which cleaves both strands of DNA (21), is involved in the terminal stages of DNA replication.
In the following, we describe the use of the murine homologue of the human WRN to create a deletion mutation in embryonic stem (ES) cells, mouse embryonic fibroblasts (MEFs), and a putative mouse model for WS. Murine ES cells and MEFs phenocopy aspects of the behavior of cells derived from human WS patients. Furthermore, biochemical studies allow us to suggest that the Werner helicase copurifies with the multiprotein DNA replication complex.
MATERIALS AND METHODS
Reverse Transcription (RT)-PCR.
Total RNA was prepared from primary cultures, cell lines, and various mouse tissues by the guanidine thiocyanate/CsCl procedure (22). Total RNA was converted to single-stranded cDNA by murine reverse transcriptase and amplified with DNA Taq polymerase (Boehringer Mannheim) as described (23). The degenerate oligonucleotides used for the PCRs are 5′-GT(A/G/C/T)ATGGC(A/G/C/T)AC(A/G/C/T)GG(A/G/C/T)GG(A/G/C/T)TA(C/T)GG-3′ for the sense primer (oligonucleotide 570) and 5′-TC(A/G/C/T)GCTTTATT(A/G/T)AT(A/G/C/T) CCCAT-3′ for the antisense primer (oligonucleotide 832). Primer 570 is based on the human amino acid sequence of motif I and primer 832 corresponds to the amino acid sequence of motif V of the DNA helicase domain of the human WS protein (12).
The analysis of mutant transcript synthesized from the disrupted allele was performed by amplifying the cDNA region coding for amino acids 652-1022 with primer Ex-A (5′-CTGGGCTCTCTTAAAACAGCGCT-3′) and primer Spe2A (5′-ACTGCAAACTGGCTTCTCCAA-3′). PCR products were separated on a 1% agarose gel and transferred on a GeneScreen membrane (DuPont) for Southern blot analysis with an internal labeled oligonucleotide as described by the manufacturer.
Screening of cDNA and Genomic DNA Libraries.
A λgt10 cDNA library, derived from a 17-day-old embryo (CLONTECH), and a NIH 3T3 cDNA library, cloned into lambda Zap (Stratagene), were used to isolate cDNA clones for murine WS’s specific mRNA. About 2 × 106 phage plaques from each library were screened by using plaque hybridization technique under stringent conditions with a partial murine WS cDNA obtained by RT-PCR. Approximately 1 × 106 phage plaques from a 129/SvEv mouse strain genomic DNA library cloned into lambda Fix II vector (Stratagene) also were screened under similar conditions. All enzymatic reactions were performed as described by the manufacturers. Standard protocols for cDNA library screening, lambda phage purification, agarose gel electrophoresis, and plasmid cloning were used (24). Northern blot analysis were performed as described (23).
PCR-SSCP.
PCR was performed in a total volume of 50 μl of water containing 0.5 mM dNTPs, 5 μl of 10× amplification buffer, 2.5 units of Taq DNA polymerase (Boehringer Mannheim), 25 ng of genomic DNA, 100 ng each of the MS-1 and MS-2 oligonucleotides, and 1 μCi of [α32-P] dCTP. PCR was carried out in a Perkin–Elmer thermal cycler through 30 cycles each of 94°C for 1 min, 55°C for 1 min, and 72°C for 2 min. The MS-1 and MS-2 oligonucleotides were designed based on the genomic sequence of an intronic region of the murine Wrn gene homologue. The MS-1 primer sequence is 5′-TTGTCGTCTACGGCATTGATC-3′ and the MS-2 primer sequence is 5′-ATACAGCTAGAATGACTCTGC-3′. These primers flank a region of genomic DNA containing a GTTTT motif repeated nine times and give a PCR product of 390 bp with C57BL/6J or Mus spretus genomic DNA. The (C57BL/6JEi × SPRET/Ei)F1 × SPRET/Ei (BSS) and the (C57BL/6J × M. spretus) F1 × C57BL/6J (BSB) DNA panels (The Jackson Laboratory) were used for the reactions and products were analyzed on a 0.5× 90 mM Tris-borate/2 mM EDTA/6% polyacrylamide gel. The gel was run at 4°C at a constant power of 40W, dried, and exposed for 1 night. Results were compiled and sent to The Jackson Laboratory for linkage analysis.
Targeting Vector.
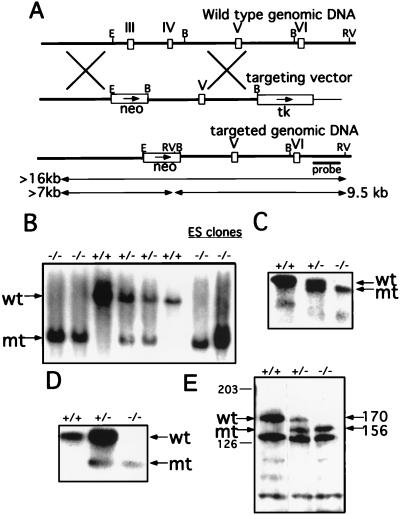
The genomic DNA of the Wrn locus from one positive recombinant phage was subcloned into the NotI site of the pBlueScript vector for restriction enzyme analysis. To construct the targeting vector, a 4-kb NotI/EcoRI fragment (the NotI site is from the recombinant phage) was subcloned into the NotI/XhoI sites of the pPNT vector (25). This construct was cleaved with BamHI and a 6-kb BamHI genomic fragment of the Wrn locus was inserted. The finished construct is shown in Fig. 2A.
Figure 2.
Targeted disruption of the helicase domain of the murine Wrn gene. (A) Schematic representations of the genomic locus surrounding the targeted exons, the targeting vector, and the targeted locus. Open boxes represent exons coding for motifs III, IV, V, and VI of the helicase domain. Transcriptional directions of the neomycin (neo) and the thymidine (tk) genes are indicated by arrows. Before electroporation, the targeting vector was linearized at the NotI site of the vector (not shown). The length of the EcoRV diagnostic restriction fragments hybridizing to the external probe are indicated by the double-headed arrows. B, BamHI; E, EcoRI; RV, EcoRV. (B) Southern blot analysis of the tail DNA from several wt, heterozygous, and homozygous mice and two homozygous ES clones. Genomic DNA was digested with EcoRV and hybridized to the probe indicated in A. The wt band is ≈16 kb and the disrupted allele (mt) generates a 9.5 kb band. (C) Northern blot analysis of the mRNA purified from the kidneys of wt, heterozygous, and homozygous mice. Five micrograms of poly(A)+ RNA was loaded in each lane. The membrane was hybridized with a labeled 3.1-kb cDNA fragment of Wrn. Arrows indicate the wt 6.6-kb full-length (wt) and mutant (mt) transcripts. (D) RT-PCR analysis of purified total RNA from the spleen of wt, heterozygous, and homozygous mice. cDNA was synthesized and used as template for amplification with the primers as described. The primer set directs the amplification of 1,116- and 753-bp fragments from the wt and mutant (mt) transcripts, respectively. PCR products were blotted and the membrane was hybridized with an internal oligonucleotide. (E) Western blot analysis of WS protein expression in wt, heterozygous, and homozygous ES clones. Arrows on left indicate the wt and the mutant (mt) proteins detected with the anti-WS helicase rabbit antibody. Arrows on right indicates the approximate molecular mass of the wt (170 kDa) and mutant (156 kDa) proteins.
Homologous Recombination in ES Cells and Generation of Germ-Line Chimeras.
TC1 ES cells were transfected with the NotI-digested targeting vector and selected with G418 and 1-(2-deoxy-2-fluoro-β-d-arabinofuranosyl)-5-iodouracil. The culture, electroporation, and selection of ES cells were carried out as described (25). The resistant ES cell colonies were picked and analyzed by Southern blotting for homologous recombination events within the Wrn locus. Genomic DNAs from these clones were digested with Eco-RV followed by the Southern transfer analysis by using a 1.2-kb HindIII/NotI fragment 3′ to the targeting vector (see Fig. 2A).
ES cells heterozygous for the targeted mutation were microinjected into C57BL/6 blastocysts to obtain germ-line transmission as described (25). Male chimeras (identified by the presence of agouti coat color) were mated with NIH Black Swiss (Taconic Farms) and with 129/SvEv females. Germ-line transmission was confirmed by Southern blot analysis.
Cell Culture and Treatments.
Generation and maintenance of the embryonic fibroblasts has been described (25). The genotype for each cell line was verified by Southern blot analysis. Cell proliferation and maximal density were determined by plating 5 × 104 cells in 6-well plates. The cultures were maintained for up to 11 days with changing media every other day. Cells were counted daily with a hemocytometer.
To obtain homozygous ES cells, heterozygous cells were incubated with higher concentrations of G418 (up to 1.2 mg/ml) and surviving colonies were picked and expanded for Southern blot analysis (26). Clones that did not show recombination at the second allele after such treatment were kept as heterozygous representatives. Likewise, ES cells that showed no homologous recombination at the Wrn locus were kept for genotype comparison in the following experiments. The sensitivity of ES cells to increasing doses of DNA-damaging agents and topoisomerase inhibitors (Sigma) was determined by measuring their colony-forming ability. Approximately 5,000 ES cells were plated onto 6-well plates containing feeder layers. After 24 hr, cells were incubated for 1 hr with either mitomycin C, 9,10-dimethyl-1,2-benz-anthracene (DMBA), methyl methane thiosulfonate, etoposide, camptothecin, or 6-thioguanine. The cells then were washed twice with PBS and allowed to grow in fresh media for 7 days. Ionizing radiation sensitivity was determined by comparing the colony-forming ability of ES cells after irradiation with an 137Cs source. For UV radiation, ES cells were plated on a feeder layer, and 24 hr later, cells were washed with PBS and exposed to a UV source. Cells were then grown for 7 days, fixed, and the colonies were counted.
Protein Analysis.
Total cell protein (50 μg) from the indicated cells was prepared as described (27) and subjected to SDS/PAGE on an 8% gel. Proteins were transferred to nitrocellulose and probed as described (27) with a rabbit polyclonal antibody (Covance Research Products, Denver, PA) raised against a glutathione S-transferase fusion protein containing the first 42 aa residues of the N-terminus portion of Wrn protein.
Fractionation Procedure.
ES cells were fractionated as described by Wu et al. (28). Five grams of frozen pelleted cells were thawed and resuspended in 3 vol of buffer containing 50 mM Tris⋅HCl (pH 7.5), 0.25 M sucrose, 5 mM MgCl2, 0.1 mM phenylmethylsulfonyl fluoride (PMSF), and 1 mM DTT. The resuspended cells were homogenized with a loose-fitting Dounce homogenizer. The homogenate then was subjected to a series of centrifugation steps to obtain postmicrosomal and nuclear fractions.
The crude nuclear extract was resuspended in 2 vol of a buffer containing 50 mM Tris⋅HCl (pH 7.5), 0.15 M KCl, 5 mM each of EDTA-Na3 and EGTA-Na3, 0.1 mM PMSF, and 1 mM DTT and gently stirred for 2 hr at 4°C. The extracted nuclei were centrifuged at 100,000 × g for 60 min and the supernatant was collected. The latter supernatant and the postmicrosomal supernatant were pooled and made 2 M in KCl. Polyethylene glycol (PEG) was added to a final concentration of 5% and the mixture stirred gently for 1 hr at 4°C. PEG-precipitated material was pelleted by centrifugation for 30 min at 16,000 × g and the supernatant was dialyzed for 3 hr as described (28). The clarified fraction was layered over a 2 M sucrose cushion and subjected to centrifugation at 100,000 × g for 18 hr at 4°C. The material above the sucrose interphase was collected and designated the HS-4 fraction. The sucrose interphase fraction was collected and designated HSP-4. The HSP-4 fraction was dialyzed and the dialyzate was chromatographed onto a column of DE52 cellulose as described (28).
RESULTS
Cloning and Mapping the Mouse Wrn Gene.
To isolate a cDNA corresponding to the mouse homologue of the human WRN gene (12), degenerate oligonucleotides were used in conjunction with RT-PCR to generate probes from murine mRNAs. These probes were used in turn to screen murine cDNA libraries (as described in Materials and Methods). A sequence of overlapping cDNA clones revealed a long ORF encoding 1,400 aa residues (data not shown). Comparison of this sequence to that of the human WS protein revealed a homology that was greater than 70% as described by Imamura et al. (29).
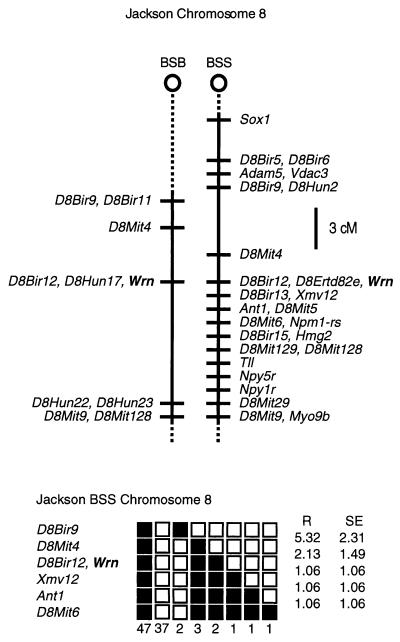
Although sequence homologies strongly support the notion that the murine gene is a homologue of the human WRN gene, we wanted to confirm this by asking whether the two genes map to syntenic regions of the human and mouse genomes. Accordingly, the interspecific backcross panels BSB and BSS were obtained from The Jackson Laboratory. These maps are anchored by a large number of previously mapped loci (30). Although the details of this analysis were given previously, briefly put, a polymorphic allele (based on a simple sequence repeat located in a Wrn intron) detected by PCR-SSCP distinguished between C57BL/6J and M. spretus alleles and could be scored for chromosomal mapping. The data were analyzed for linkage by The Jackson Laboratory and it was found that the Wrn gene cosegregated with an anonymous DNA marker (D8Bir12) located on mouse chromosome 8 (Fig. 1), consistent with the fluorescence in situ hybridization assignment of Imamura et al. (29) and syntenic to the human WRN gene on human 8p (31, 32). Raw data from The Jackson Laboratory were obtained from the World Wide Web address http://www.jax.org/resources/documents/cmdata. Thus, the syntenic chromosomal location of mouse Wrn and human WRN genes and their tight sequence homologies strongly support the conclusion that these genes are homologues of one another.
Figure 1.
Mapping of the murine Wrn gene. (Upper) Maps from the Jackson BSB and BSS backcrosses showing part of chromosome 8. Maps are depicted with centromere toward the top. A 3-centimorgan scale is shown at right. Loci mappings to the same position are listed alphabetically. Missing typing was inferred from surrounding data, where assignment was unambiguous. (Lower) Haplotype figure from the Jackson BSS backcross showing part of chromosome 8 with loci linked to Wrn. Loci are listed in order with the most proximal at the top. Solid boxes represent the C57BL6/JEi allele and open boxes the SPRET/Ei allele. The number of the animals with each haplotype is given at the bottom of each column of boxes. The percent recombination (R) between adjacent loci is given to the right with the standard error. Missing typing was inferred from surrounding data, where assignment was unambiguous.
Generation of Mutant Mice Bearing a Deletion of a Portion of the WS Helicase Domain.
Like other members of the helicase superfamily, the human WRN gene encodes a central motif composed of seven domains constituting the helicase signature. Several WS patients have been reported to carry WS mutations involving these domains, including the specific deletion of exons encoding helicase domains III and IV (13). The mouse WS protein also contains these seven helicase domains, and domains III and IV are encoded by two separate exons as in the human gene homologue (13). To recapitulate part of this human mutation, we designed a homologous recombination construct to replace a 4.1-kb genomic fragment containing these exons with a neo cassette (Fig. 2A). Transcription of this mutated gene could give rise to two differently processed mutant WS RNA transcripts; one including the neo cassette that would result in a truncated WS protein, and a second, in which the neo cassette would be removed by RNA splicing and from which a mutant WS protein deleted of helicase domains III and IV would be produced. Both mutant forms should lack helicase activity.
The mutation shown in Fig. 2A was produced by homologous recombination in 129/SvEv ES cells (TC1) (25), which were in turn introduced into blastocysts to create chimeric mice using standard techniques as described. Mice of all possible genotypes were generated by mating the chimeras with either 129/SvEv or NIH Black Swiss mice and intercrossing the F1 generation. All three expected genotypes were detected (Fig. 2B). The ratio of wt/heterozygote/homozygote mice in the NIH Black Swiss outbred background falls short of the expected 1:2:1 Mendelian ratio (actual, 1:2:0.8) among a total of 295 pups analyzed. Likewise, the ratio on the 129/SvEv inbred background also indicates that the number of homozygous WS mice is lower than expected (1:1.9:0.6) in a total of 154 pups analyzed. Although these results indicate that homozygous WS are viable (as are human WS patients), the distortion of the Mendelian ratio suggests that the homozygote state confers a survival disadvantage, a possibility that has not as yet been evaluated in humans.
The interrupted Wrn gene could result in an mRNA that encodes a deletion of a segment of the helicase motif or a truncated WS protein. To determine which (if not both) of the two possible WS RNA transcripts are produced by the disrupted Wrn gene, a Northern blot analysis of kidney poly(A)+ RNA derived from wt, heterozygous, and homozygous mice was performed. As shown in Fig. 2C, a 6.6-kb transcript was detected in both wt and heterozygous samples. By contrast, this transcript was absent from the homozygous mutant mouse sample, which displayed a transcript of somewhat lower molecular weight (Fig. 2C). The same transcript was also detected in the heterozygous sample and formed a doublet with the wt transcript (Fig. 2C, middle lane). Both the wt and the mutant transcripts were of equal intensity in the heterozygous animals. Consistent with a mechanism by which the neo insert is spliced out of the RNA transcript of the mutant gene, a neo probe failed to hybridize to either of the transcripts. In addition, no transcript larger than 6.6 kb could be detected with either the WS or the neo probes (data not shown).
The mutant transcript was characterized further by using RT-PCR analysis of the mRNA region encoding a major portion of the WS helicase domain. As indicated in Fig. 2D, the sample from the wt mouse showed a single amplification product corresponding to the portion of the helicase domain analyzed. The heterozygous sample yielded two bands, one corresponding to the wt allele and another, 363 bp shorter, corresponding to the mutant allele. As expected, the homozygous mutant sample yielded only the smaller amplification product. This smaller RT-PCR fragment was cloned and sequenced, confirming the 363-nt deletion and confirming that two exons have been removed by homologous recombination. The neo cassette is removed at the level of mRNA splicing. Furthermore, this structural analysis indicates that translation of the mutant transcript should direct the synthesis of a mutant WS protein lacking the 121 aa corresponding to the exons deleted from the helicase domain.
A Smaller WS Protein Is Synthesized from the Mutated Allele.
To analyze both mutant and wt WS proteins, antibodies were raised against the N-terminal portion (the first 42 aa) of the murine WS protein. Because of the high level of expression of WS mRNA in ES cells, extracts of these cells were used for the initial protein analyses. The homozygous ES cells were created by selecting G418 resistant cells from among heterozygous ES cells cultured at increasing concentrations of G418. The genotype of each clone was confirmed by DNA blot analysis (Fig. 2B).
Total cell proteins were extracted from the wt, heterozygous, and homozygous ES cells for protein blot analysis. As shown in Fig. 2E, the polyclonal antibody recognizes a protein of ≈170 kDa consistent with the mass expected for the wt WS protein. The level of expression of the 170-kDa protein in heterozygous cells is approximately one-half the level of expression of the same protein in homozygous cells. In addition, a protein of 156 kDa is also detected in the heterozygous cells and is of equal amount to the 170-kDa protein in that cell line. The mass of the mutant protein corresponds to the expected loss of 121 aa. Also, as expected, only the mutant protein is detected in homozygous ES cells (Fig. 2E). These results indicate that the mutant gene directs the synthesis of a mutant WS protein and, moreover, that the mutant protein is relatively stable.
Phenotype of the Mutant Mice.
As indicated above, the reduced Mendelian ratio of surviving WS homozygotes suggests an embryonic or perinatal survival disadvantage. While those surviving homozygotes appear to grow normally, one of the two males autopsied at 10 months of age displayed extensive myocardial fibrosis not seen in age/sex matched heterozygous littermates or wt controls (not shown). In addition, the oldest homozygous female (13.5 months old) developed a T cell lymphoma, whereas none of the 17 control (the same age or older) heterozygous littermates have thus far developed any malignancies. Most homozygous mice have been observed for fewer than 13 months, largely in an outbred genetic background. Obviously, firm conclusions regarding their longevity and further age-related morbidity await a longer period of observation and an assessment of the potential effects of genetic background.
6-Thioguanine-Resistant Colonies Arise More Frequently from WS ES Cells Than From wt, Although Several DNA Repair Pathways Remain Intact.
The availability of homozygous Wrn-deficient ES cells provided the opportunity to test a surrogate for spontaneous mutagenesis and clonogenic survival, the latter as a function of DNA damage induced by a variety of treatments. Growth curve analyses of all the ES clones indicated that mutant ES cells did not differ in their proliferative potential from wt cells, even after several passages (data not shown). Moreover, the plating efficiencies (10–20%) were virtually identical for both mutant and wt ES cells.
Given the above-noted properties, the spontaneous mutation rates of the ES cells were examined by using the surrogate property of resistance selection against the antileukemic agent 6-thioguanine (33, 34). This purine analogue induces its lethal effect as a result of its incorporation into DNA. However, for 6-thioguanine to be incorporated into DNA, it requires the action of the X-linked enzyme hypoxanthine guanine phosphoribosyltransferase (HPRT). Any spontaneous mutations inactivating the HPRT locus will render cells resistant to 6-thioguanine. Thus, male-derived ES cells were treated with increasing concentrations of 6-thioguanine and resistant colonies were scored. As shown in Fig. 3A, at the highest concentration of the drug tested, the average frequency of 6-thioguanine-resistant colonies was 6-fold higher in WS ES cells than in either wt or heterozygous cells. These results suggest that WS ES cells have a higher spontaneous mutation rate at least at the HPRT locus.
Figure 3.
Mutagenicity and differential effects of DNA damaging agents on wt and WS ES cells. Approximately 5,000 ES cells were plated onto a feeder layer and plates were treated for 2 hr with the indicated chemicals or irradiated, and then cells were grown for 7 days to form individual colonies. Each curve represents two independent clones plated in duplicate. Graphs represent the clonogenic survival of wt Wrn+/+, heterozygous Wrn+/−, and homozygous Wrn−/− ES cells after treatment with increasing concentrations of 6-thioguanine (A), type II topoisomerase inhibitor etoposide (B), topoisomerase I inhibitor camptothecin (C), increasing doses of γ-rays (D), increasing doses of UV light (E), and increasing concentration of mitomycin C (F).
Given the higher mutation rate in WS ES cells, different DNA damaging treatments were used to detect any repair defects that might be associated with the mutation in the WS protein. Ionizing γ-radiation introduces double-strand breaks in DNA. As shown in Fig. 3D, there is no significant difference among the different genotypic ES cells with respect to survival as a function of increasing γ-radiation. UV light damages DNA through the induction of thymine dimers that are repaired primarily by a nucleotide excision repair pathway. Again, WS ES cells were not more sensitive to UV light than their wt or heterozygous counterparts (Fig. 3E). Finally, ES cells were treated with mitomycin C, which introduces intrastrand cross-linkage in DNA (35). Again, no difference was detected among the three genotypes (Fig. 3F). Similarly, sensitivity to the alkylating agent methylmethane thio-sulfonate and the DMBA agent, which covalently binds DNA, was not increased in homozygous ES cells as compared with either their wt or heterozygous counterparts (data not shown).
Homozygous Mutant ES Cells Display an Increased Sensitivity to Inhibitors of Topoisomerase.
The Wrn gene product is homologous to the yeast DNA helicase Sgs1p (17), a protein that interacts with topoisomerase II and is required for faithful chromosome segregation (16). The yeast DNA helicase also can interact with the yeast type I topoisomerase (15). Interestingly, increased concentrations of topoisomerases can be found in certain malignancies, which in turn can be treated by antitumor drugs that act as topoisomerase inhibitors (36, 37). These drugs stabilize the DNA/topoisomerase cleavage complex, resulting in DNA strand breaks that may not be religated or repaired. To test for a possible interaction between topoisomerases and the WS helicase, ES cells of all three genotypes were treated with the type I topoisomerase inhibitor, camptothecin (37, 38), and the topoisomerase type II inhibitor, etoposide (36). As shown in Fig. 3B, homozygous WS ES cells were 2- to 3-fold more sensitive to etoposide than were heterozygous or wt ES cells. The same homozygous ES cells were 10 times more sensitive to increasing concentrations of camptothecin than the wt ES cells (Fig. 3C), whereas the heterozygous cells demonstrated an intermediate level of sensitivity. These results suggest that there is a biochemical interaction between the WS helicase and one (or more) of the topoisomerases.
Homozygous WS MEFs Display a Prematurely Reduced Growth Rate as a Function of Passage in Culture.
Because reduced growth rate and premature senescence are properties generally associated with the premature aging of human WS fibroblasts (3), we wanted to examine this property in fibroblasts derived from the mouse model. Thus, MEFs were established from 17-day-old embryos produced from a mating of heterozygous (Wrn+/−) mice. After genotyping the primary cultures, the growth potential of the cells was analyzed as a function of passage. During the first three passages no differences were noted in the growth rates. It was only after the fifth passage that differences could be observed. As indicated in Fig. 4, WS MEFs grow more slowly and are saturated at lower densities in culture than are their heterozygous and wt counterparts.
Figure 4.
Differential saturation density and growth properties of wt and WS MEFs. Saturation density and growth analysis of MEFs at passage 5. Cells (5 × 104) from Wrn+/+, Wrn+/−, and Wrn−/− were plated in 6-well plates as described. Cell number at each time point represents the average of two independent cell lines.
The WS Protein Copurifies with the Multiprotein DNA Replication Complex.
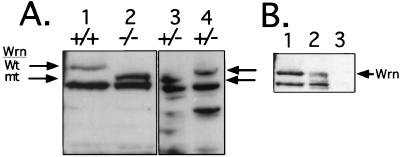
The effect of camptothecin suggests that the WS helicase acts in concert with topoisomerases. This potential interaction could take place during DNA replication. Indeed, topoisomerase I has been shown to be part of the DNA replication apparatus (28, 39) and the topoisomerase I inhibitor, camptothecin, induces double-strand breaks in newly replicated DNA. Such results suggest that the WS DNA helicase might be a part of the DNA replication apparatus whether or not it is involved in replication, repair, or some other aspect of the activity of this complex. Accordingly, analyses were performed to determine whether the mutant and the wt proteins are found in cellular fractions containing the well-characterized multiprotein DNA replication complex. As shown in Fig. 2E, our antibody readily detects both mutant and wt WS proteins that are expressed at similar levels in heterozygous ES cells.
To demonstrate this potential association, fractionation of the WS ES cell proteins was performed according to a protocol that results in the substantial purification of multiprotein DNA replication complex. Briefly, extracted proteins were incubated with PEG and the protein complexes centrifuged onto a 2 M sucrose cushion and divided into an interphase fraction consisting of high molecular weight protein complexes (including the multiprotein DNA replication complex) (fraction HSP-4) and a supernatant fraction consisting of soluble proteins (fraction HS-4). Fig. 5A shows that the wt WS protein is found in the HSP-4 complex fraction (lane 4), but not in the HS-4 (supernatant) fraction (lane 3), suggesting that the WS protein is part of a multiprotein complex. Previous studies had indicated that helicase activity can indeed be detected in this HSP-4 complex fraction, but not in the soluble HS-4 fraction (28). Most importantly, however, we see that the mutant WS protein is not associated with the complex fraction, but can now be detected among the soluble proteins in the supernatant fraction (Fig. 5A, lane 3).
Figure 5.
Copurification of WS protein and the multiprotein DNA replication complex is disrupted by the Wrn mutation. (A) Total cell lysates (50 μg) from wt (+/+) (lane 1) and homozygous (−/−) ES cells (lane 2) were loaded on the gel as reference. Lanes 3 and 4 contain protein from heterozygous (+/−) ES cells fractionated on a sucrose cushion as described. Lane 3, protein from the HS-4 supernatant fraction; lane 4, protein from the HSP-4 interphase fraction containing protein complexes. Proteins were size separated on 10% SDS/PAGE and detected by Western blot analysis with the anti-WS antibody. Arrows indicate the wt and the mutant (mt) WS proteins. Lanes 3 and 4 from the same autoradiogram were exposed for longer than lanes 1 and 2. (B) Immunoblot analysis for the presence of WS protein in DE52 low and high salt eluates. Lanes: 1, total cell lysate from wt ES cells; 2, protein (20 μg) from the high salt DE52 eluate; 3, protein (20 μg) from the DE52 wash fraction. Arrow indicates the wt protein.
As described previously (28), the HSP-4 complex fraction contains the bulk of DNA polymerase activity and thus the multiprotein DNA replication complex (data not shown). To extend this analysis, the multiprotein DNA replication complex was purified further by DE52 chromatography. Proteins were loaded in low salt buffer and the bound proteins were eluted with a high salt buffer. As shown in Fig. 5B (lane 2), the wt WS protein is detected in the high salt eluate. Again, previous studies have shown that helicase activity, in addition to other components of the multiprotein DNA replication complex, are associated with the high salt eluate (28). These data indicate that the wt WS protein copurifies with the multiprotein DNA replication complex and, therefore, might be a part of that complex. Moreover, the possibility that this association is a physiological one is suggested by the fact that mutant WS protein fails to associate with the complex (Fig. 5A, lane 3).
DISCUSSION
Given the similarity of the human and murine genes, we wanted to create a mutation that resembled those seen in the human WS. Some of these mutations have been described as deletions of one or several exons coding for part of the helicase domain of the protein (13). Accordingly, we deleted motifs III and IV of the helicase domain, creating a mutation that could result in the deletion of a major segment of the helicase signature or a truncated WS protein. Our detailed studies involving transcript sequencing and protein characterization indicate that the mutant gene produces a stable version of the WS protein, which is deleted with respect to a major portion of the helicase domain. The mutant gene is widely expressed as is the wt gene and, as in the human, homozygous deficient mice are viable.
Although it will take time to evaluate the long-term effects of the murine Wrn mutation, several aspects can be examined and compared at the cellular level. We have noted that late-passage WS mouse fibroblasts show reduced proliferation and early saturation arrest as compared with wt fibroblasts, a behavior that is seen in fibroblasts from WS patients and older individuals (3). In addition, it has been reported that the spontaneous mutation rate at the human HPRT locus is elevated in immortalized cell lines derived from WS patients as compared with cells derived from normal donors (40–42). We have carried out a similar experiment using murine WS ES cells challenged with the purine analogue, 6-thioguanine, and have found that homozygous murine ES cells also have a higher rate of induction of resistant colonies as compared with wt cells.
In addition to assessments of mutation rate, there have been extensive studies directed toward examining the possibility that the WS helicase plays a role in one or more of the DNA repair pathways. Studies on human WS lymphocytes and fibroblasts fail to show any increased sensitivity in a variety of DNA damage repair assays (43–45). Similarly, we have evaluated the response of our mutant WS ES cells to a number of DNA damaging agents. Our data indicate that WS ES cells are not any more sensitive than wt cells to either γ-irradiation, UV light, 4–6 cyclobutane pyrimidines, or mitomycin C. Similarly, mutant ES cells do not show an increased sensitivity to the mutagen DMBA or the alkylating agent methyl methanesulfonate.
We have already noted that the WS helicase bears sequence similarity to the helicase domain of the S. cerevisiae gene product, Sgs1p (17). Furthermore, the Sgs1 helicase can interact with type I and II topoisomerases in yeast cells (15, 16). It is thus reasonable to ask whether the mutant WS protein might alter the sensitivity of WS cells to agents that block the action of one or the other topoisomerase. Indeed, it has been shown that human WS lymphoblasts show an increased sensitivity to both topoisomerase type I and II inhibitors, camptothecin and etoposide (46, 47). Consistent with this, we have shown that the murine mutant ES cells are similarly more sensitive to etoposide and even more sensitive to the type I topoisomerase inhibitor, camptothecin. Since both topoisomerases are involved in DNA replication, it is possible that the WS helicase acts in concert with topoisomerase, an action that could take place during DNA replication, transcription, or recombination. Topoisomerase I has been shown to be part of the DNA replication apparatus (28, 39). The topoisomerase I inhibitor camptothecin acts mainly during S-phase, inducing toxic double-strand breaks in newly replicated DNA. This toxicity can be prevented by coexposure to the DNA synthesis inhibitor aphidicolin, indicating that ongoing DNA synthesis is required for production of double-strand breaks and cell killing (48). Because WS ES cells are more sensitive to camptothecin than wt cells, it was logical to ask whether WS protein associates with the multiprotein DNA replication complex. Our copurification data suggest that the WS protein might interact with components of this replication complex. Further biochemical characterization will be necessary to establish this point.
Acknowledgments
We are grateful to C. Daugherty-O’Hara for helpful technical assistance. M.L. is a recipient of a National Cancer Institute of Canada Research Fellowship supported with funds provided by the Terry Fox Run.
ABBREVIATIONS
- WS
Werner syndrome
- wt
wild type
- RT
reverse transcription
- ES
embryonic stem
- MEFs
mouse embryonic fibroblasts
- HPRT
hypoxanthine guanine phosphoribosyltransferase
- BSS
(C57BL/6JEi × SPRET/Ei)F1 × SPRET/Ei
- BSB
(C57BL/6J × M. spretus) F1 × C57BL/6J
Footnotes
Data deposition: The mouse Wrn mapping data reported in this paper has been deposited in the Mouse Genome Database (accession no. J: 48424).
References
- 1. Epstein C J, Martin G M, Schultz A L, Motulsky A G. Medicine. 1966;45:177–222. doi: 10.1097/00005792-196605000-00001. [DOI] [PubMed] [Google Scholar]
- 2.Sato K, Goto M, Nishioka K, Arimu K, Hori N, Yamashita N, Fujimoto Y, Nanko H, Ohara K. Gerontology. 1988;34:212–218. doi: 10.1159/000212956. [DOI] [PubMed] [Google Scholar]
- 3.Salk D, Bryant E, Au K, Hoehn H, Martin G M. Hum Genet. 1981;58:310–316. doi: 10.1007/BF00294930. [DOI] [PubMed] [Google Scholar]
- 4.Faragher R G A, Kill I R, Hunter J A A, Pope F M, Tannock C, Shall S. Proc Natl Acad Sci USA. 1993;90:12030–12034. doi: 10.1073/pnas.90.24.12030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schulz V P, Zakian V A, Ogburn C E, McKay J, Jarzebowicz A A, Edland S D, Martin G M. Hum Genet. 1996;97:750–754. doi: 10.1007/BF02346184. [DOI] [PubMed] [Google Scholar]
- 6.Schonberg S, Niermeijer M F, Bootsma D, Henderson E, German J. Am J Hum Genet. 1984;36:387–397. [PMC free article] [PubMed] [Google Scholar]
- 7.Scappaticci S, Forabosco A, Borroni G, Orecchia G, Fraccaro M. Ann Genet. 1990;33:5–8. [PubMed] [Google Scholar]
- 8.Goto M, Rubenstien M, Weber J, Woods K, Dryana D. Nature (London) 1992;355:735–738. doi: 10.1038/355735a0. [DOI] [PubMed] [Google Scholar]
- 9.Schellenberg G D, Martin G M, Wijsman E M, Nakura J, Miki T, Ogihara T. Lancet. 1992;339:1002. doi: 10.1016/0140-6736(92)91590-5. [DOI] [PubMed] [Google Scholar]
- 10.Goddard K A B, Yu C-E, Oshima J, Miki T, Nakura J, Piussan C, Martin G M, Schellenberg G D, Wijsman E M The International Werner’s Syndrome Collaborative Group. Am J Hum Genet. 1996;58:1286–1302. [PMC free article] [PubMed] [Google Scholar]
- 11.Nakura J, Miki T, Ye L, Mitsuda N, Zhao Y, Kihara K, Yu C-E, Oshima J, Fukuchi K-I, Wijsman E M, et al. Genomics. 1996;36:130–141. doi: 10.1006/geno.1996.0433. [DOI] [PubMed] [Google Scholar]
- 12.Yu C-E, Oshima J, Fu Y-H, Wijsman E M, Hisama F, Alish R, Matthews S, Nakura J, Miki T, Quais S, et al. Science. 1996;272:258–262. doi: 10.1126/science.272.5259.258. [DOI] [PubMed] [Google Scholar]
- 13.Yu C-E, Oshima J, Wijsman E M, Nakura J, Miki T, Piussan C, Matthews S, Fu Y-H, Mulligan J, Martin G M, et al. Am J Hum Genet. 1997;60:330–341. [PMC free article] [PubMed] [Google Scholar]
- 14.Umezu K, Nakayama K, Nakayama H. Proc Natl Acad Sci USA. 1990;87:5363–5367. doi: 10.1073/pnas.87.14.5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gangloff S, McDonald J P, Bendixen C, Arthur L, Rothstein R. Mol Cell Biol. 1994;14:8391–8398. doi: 10.1128/mcb.14.12.8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Watt P M, Louis E J, Borts R H, Hickson I D. Cell. 1995;81:253–260. doi: 10.1016/0092-8674(95)90335-6. [DOI] [PubMed] [Google Scholar]
- 17.Watt P M, Hickson I D, Borts R H, Louis E J. Genetics. 1996;144:935–945. doi: 10.1093/genetics/144.3.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kretzschmar M, Meisterernst M, Roeder R G. Proc Natl Acad Sci USA. 1993;90:11508–11512. doi: 10.1073/pnas.90.24.11508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang H, Wang J C, Liu L F. Proc Natl Acad Sci USA. 1988;85:1060–1064. doi: 10.1073/pnas.85.4.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sinclair D A, Mills K, Guarente L. Science. 1997;277:1313–1316. doi: 10.1126/science.277.5330.1313. [DOI] [PubMed] [Google Scholar]
- 21.Kallio M, Lähdetie J. Mutagenesis. 1996;11:435–443. doi: 10.1093/mutage/11.5.435. [DOI] [PubMed] [Google Scholar]
- 22.Chirgwin J M, Przybyla A E, MacDonald R J, Rutter W J. Biochemistry. 1979;13:2633–2637. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- 23.Lebel M, Mes-Masson A-M. Exp Cell Res. 1994;213:12–19. doi: 10.1006/excr.1994.1167. [DOI] [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 25.Deng C, Zhang P, Harper J W, Elledge S J, Leder P. Cell. 1995;82:675–684. doi: 10.1016/0092-8674(95)90039-x. [DOI] [PubMed] [Google Scholar]
- 26.Mortensen R M, Conner D A, Chao S, Geisterfer-Lowrance A A T, Seidman J G. Mol Cell Biol. 1992;12:2391–2395. doi: 10.1128/mcb.12.5.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harlow E, Lane D. Antibody: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. [Google Scholar]
- 28.Wu Y, Hickey R, Lawlor K, Wills P, Yu F, Ozer H, Starr R, Quan J Y, Lee M, Malkas L. J Cell Biochem. 1994;54:32–46. doi: 10.1002/jcb.240540105. [DOI] [PubMed] [Google Scholar]
- 29.Imamura O, Ichikawa K, Yamabe Y, Goto M, Sugawara M, Furuichi Y. Genomics. 1997;41:298–300. doi: 10.1006/geno.1997.4661. [DOI] [PubMed] [Google Scholar]
- 30.Rowe L B, Nadeau J H, Turner R, Frankel W N, Letts V A, Eppig J T, Ko M S H, Thurston S J, Birkenmeier E H. Mamm Genome. 1994;5:253–274. doi: 10.1007/BF00389540. [DOI] [PubMed] [Google Scholar]
- 31.Copeland N G, Jenkins N A, Gilbert D J, Eppig J T, Maltais L J, Miller J C, Dietrich W F, Weaver A, Lincoln S E, Steen R G, et al. Science. 1993;262:57–66. doi: 10.1126/science.8211130. [DOI] [PubMed] [Google Scholar]
- 32.Lyon M F, Cocking Y, Gao X. Mouse Genome. 1996;94:29–73. [Google Scholar]
- 33.Maybaum J, Bainnson A N, Roethel W M, Ajmera S, Iwaniec L M, TerBush D R, Kroll J J. Mol Pharmacol. 1987;32:606–614. [PubMed] [Google Scholar]
- 34.Uribe-Luna S, Quintana-Hau J D, Maldonado-Rodriguez R, Espinosa-Lara M, Beattie K L, Farquhar D, Nelson J A. Biochem Pharmacol. 1997;54:419–424. doi: 10.1016/s0006-2952(97)00200-1. [DOI] [PubMed] [Google Scholar]
- 35.Tomasz M, Lipman R, Chowdary D, Pawlak J, Verdine G, Nakanishi K. Science. 1987;235:1204–1208. doi: 10.1126/science.3103215. [DOI] [PubMed] [Google Scholar]
- 36.Wang J C. Annu Rev Biochem. 1985;54:665–697. doi: 10.1146/annurev.bi.54.070185.003313. [DOI] [PubMed] [Google Scholar]
- 37.Giovanella B C, Stehlin J S, Wall M E, Wani M C, Nicholas A W, Liu L F, Silber R, Potmesil M. Science. 1989;246:1046–1048. doi: 10.1126/science.2555920. [DOI] [PubMed] [Google Scholar]
- 38.Morham S G, Kluckman K D, Voulomanos N, Smithies O. Mol Cell Biol. 1996;16:6804–6809. doi: 10.1128/mcb.16.12.6804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simbulan-Rosenthal C M, Rosenthal D S, Hilz H, Hickey R, Malkas L, Applegren N, Wu Y, Bers G, Smulson M E. Biochemistry. 1996;35:11622–11633. doi: 10.1021/bi953010z. [DOI] [PubMed] [Google Scholar]
- 40.Fukuchi K, Tanaka K, Nakura J, Kumahara Y, Uchida T, Okada Y. Somatic Cell Mol Genet. 1985;11:303–308. doi: 10.1007/BF01534688. [DOI] [PubMed] [Google Scholar]
- 41.Fukuchi K, Martin G M, Monnat R J., Jr Proc Natl Acad Sci USA. 1989;86:5893–5897. doi: 10.1073/pnas.86.15.5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fukuchi K, Tanaka K, Kumahara Y, Marumo K, Pride M B, Martin G M, Monnat R J., Jr Hum Genet. 1990;84:249–252. doi: 10.1007/BF00200569. [DOI] [PubMed] [Google Scholar]
- 43.Stefanini M, Scappaticci S, Largomarsini P, Berardesca E, Nuzzo F. Mutat Res. 1989;219:179–185. doi: 10.1016/0921-8734(89)90013-1. [DOI] [PubMed] [Google Scholar]
- 44.Saito H, Moses R E. Exp Cell Res. 1991;192:373–379. doi: 10.1016/0014-4827(91)90054-x. [DOI] [PubMed] [Google Scholar]
- 45.Weirch-Schwaiger H, Weirich H G, Gruber B, Schweiger M, Hirsh-Kauffmann M. Mutat Res. 1994;316:37–48. doi: 10.1016/0921-8734(94)90006-x. [DOI] [PubMed] [Google Scholar]
- 46.Elli R, Chessa L, Antonelli A, Petrinelli P, Ambra R, Marcucci L. Cancer Genet Cytogenet. 1996;87:112–116. doi: 10.1016/0165-4608(95)00294-4. [DOI] [PubMed] [Google Scholar]
- 47.Okada M, Goto M, Furuichi Y, Sugimoto M. Biol Pharmacol Bull. 1998;21:235–239. doi: 10.1248/bpb.21.235. [DOI] [PubMed] [Google Scholar]
- 48.Ryan A J S, Squires S, Strutt H L, Johnson R T. Nucleic Acids Res. 1991;19:3295–3300. doi: 10.1093/nar/19.12.3295. [DOI] [PMC free article] [PubMed] [Google Scholar]