Abstract
Pallido-ponto-nigral degeneration (PPND) is one of the most well characterized familial neurodegenerative disorders linked to chromosome 17q21–22. These hereditary disorders are known collectively as frontotemporal dementia (FTD) and parkinsonism linked to chromosome 17 (FTDP-17). Although the clinical features and associated regional variations in the neuronal loss observed in different FTDP-17 kindreds are diverse, the diagnostic lesions of FTDP-17 brains are tau-rich filaments in the cytoplasm of specific subpopulations of neurons and glial cells. The microtubule associated protein (tau) gene is located on chromosome 17q21–22. For these reasons, we investigated the possibility that PPND and other FTDP-17 syndromes might be caused by mutations in the tau gene. Two missense mutations in exon 10 of the tau gene that segregate with disease, Asn279Lys in the PPND kindred and Pro301Leu in four other FTDP-17 kindreds, were found. A third mutation was found in the intron adjacent to the 3′ splice site of exon 10 in patients from another FTDP-17 family. Transcripts that contain exon 10 encode tau isoforms with four microtubule (MT)-binding repeats (4Rtau) as opposed to tau isoforms with three MT-binding repeats (3Rtau). The insoluble tau aggregates isolated from brains of patients with each mutation were analyzed by immunoblotting using tau-specific antibodies. For each of three mutations, abnormal tau with an apparent Mr of 64 and 69 was observed. The dephosphorylated material comigrated with tau isoforms containing exon 10 having four MT-binding repeats but not with 3Rtau. Thus, the brains of patients with both the missense mutations and the splice junction mutation contain aggregates of insoluble 4Rtau in filamentous inclusions, which may lead to neurodegeneration.
Keywords: frontotemporal dementia and Parkinsonism/tauopathies/glial and neuronal tangles/microtubule associated proteins/Alzheimer’s disease
Familial frontotemporal dementia (FTD) with parkinsonism is a group of neurodegenerative syndromes with diverse but overlapping clinical and neuropathologic features. Genetic linkage studies show that, in most of these kindreds, genetic markers from 17q21–22 cosegregate with the disease. Hence, this group of syndromes collectively is referred to as frontotemporal dementia and parkinsonism linked to chromosome 17 (FTDP-17) (1). The first two families genetically mapped to 17q21–22 were called disinhibition–dementia–parkinsonism–amyotrophy complex (DDPAC) (2) and pallido-ponto-nigral degeneration (PPND) (3). Subsequently other families with FTD were mapped definitively to 17q21–22, and the interval between genetic markers D17S800 and D17S791 was established as the critical region (1). The partial overlap in the phenotype of these kindreds combined with the common genetic location of the disease locus suggests that these heterogeneous syndromes are allelic.
In each FTDP-17 kindred, the presenting symptoms vary but reflect specific patterns of neuronal loss in the postmortem brains (1). For example, PPND patients typically present with parkinsonism alone or with parkinsonism and personality changes, correlating with the extent of neuron loss in subcortical nuclei and frontal cortex (4). In contrast, the most common initial symptom in DDPAC and FTD kindreds is a personality change reflecting the preferential degeneration of neurons in the superficial frontal cortex (1). Despite this phenotypic heterogeneity, a common feature of FTDP-17 is numerous neuronal and glial inclusions of aggregated filaments of hyperphosphorylated tau proteins associated with neuronal loss.
No pathogenic mutations were reported in the tau gene in two earlier studies of FTDP-17 kindreds (5). Poorkaj et al. (7) recently described a missense mutation [Val337Met, numbered according to Goedert et al. (8) for the longest tau isoform] in the tau gene of affected members of one FTDP-17 kindred (the Seattle A family). This prompted us to systematically search for and find three additional mutations in the tau gene in PPND and other FTDP-17 kindreds. All of these mutations are in or near coding sequence for the C-terminal tandem imperfect repeats that contain microtubule (MT)-binding sites. Differential splicing of transcripts normally leads to production of tau isoforms with three or four MT-binding sites (9). Significantly, the biological consequence of all three mutations resulted in the formation of aggregates containing four MT-binding repeats (4Rtau) in the brains of the FTDP-17 patients from these different kindreds. Thus, we suggest that the formation of 4Rtau isoform aggregates is a pathologic consequence of the mutations in the tau gene described here and that this aggregation is responsible for at least some FTDP-17 syndromes.
MATERIALS AND METHODS
Clinical and Pathological Phenotypes of the FTDP-17 Kindreds.
The clinical and pathological features of affected members of the PPND, DDPAC, and other FTDP-17 kindreds studied here have been described extensively in previous publications (3, 10–12) and are summarized here in Table 1. The Montreal family, Seattle families D and F, and Oregon family EL have familial dementia consistent with FTD. There is only limited clinical and pathologic information available for most of the other cases that were screened for mutations in this study. We screened index cases from 33 additional families with clinical or pathologic FTD, 45 index cases with familial dementia without autopsy data, and 33 index cases of late onset familial Alzheimer’s disease (AD) for mutations as described below.
Table 1.
Families with mutations segregated with the tau gene
| Condition, family | No. of affected subjects | No. of autopsied subjects | Affected subjects sampled | Total subjects sampled | Mean onset age ±SD (n) range | Mutation |
|---|---|---|---|---|---|---|
| PPND | 36 | 13 | 17 | 308 | 43.7 ± 5.5 (33) 32–58 | Asn279Lys |
| DDPAC | 13 | 6 | 7 | 33 | 44.8 ± 9.5 (13) 27–56 | Intronic |
| FTD, Montreal | 6 | 1 | 3 | 19 | 57.2 ± 2.1 (5) 55–63 | Pro301Leu |
| FTD, Seattle D | 6 | 2 | 3 | 5 | 49.4 ± 4.8 (5) 42–57 | Pro301Leu |
| FTD, Seattle F | 14 | 1 | 5 | 8 | 61 ± 3.9 (9) 56–67 | Pro301Leu |
| FTD, Oregon EL | 4 | 0 | 2 | 3 | 64.3 ± 7.7 (4) 58–75 | Pro301Leu |
Subject DNA.
DNA samples from FTDP-17 families were prepared from all available affected, at risk subjects and spouses. Control DNA was from 95 unrelated unaffected Caucasians with an average age of 39.5 ± 11.2.
Mutation Screening.
Tau exons were sequenced automatically by purification of a 50-μl PCR using Centricon-100 concentrators (Amicon) followed by sequencing using TaqFS DNA polymerase, fluorescent dye terminators, and an ABI 373 or 310 DNA sequencer (Applied Biosystems).
Mutation and Polymorphism Assays.
For each mutation and polymorphism detected, assays were developed so that family members and normal controls could be screened. Assays for polymorphisms are described elsewhere (7). For the Asn279Lys mutation, genomic DNA was amplified with the primer pair 10F (5′-CGAGCAAGCAGCGGGTCC) and 10R (5′-GTACGACTCACACCACTTCC). When digested with the restriction enzyme MboII, the mutant allele produced 136- and 86-bp fragments whereas the normal allele remained uncut. For the intronic DDPAC mutation, genomic DNA was amplified with the primer pair 10F2 (5′-AGCAAGCAGGCGGGTCCAg-3′) and 10 R2 (5′-ATTCAAGCCACAGCACGGC-3′). Digestion of the PCR product with AflIII yielded fragments of 129, 34, and 31 bp for the normal allele and of 194 and 65 bp for the mutant allele. For the Pro301Leu mutation, genomic DNA was amplified by using primer pair 10F2/10R2. On digestion, the PCR product with MspI yielded 137- and 57-bp fragments for the normal allele, and the mutant allele was uncut.
Biochemical Analysis of Tau Proteins from the Brains of Several FTDP-17 Kindreds.
Tau proteins were extracted from the insoluble fractions of frontal cortex of two affected members of the DDPAC kindred and one affected member of the Montreal kindred, and frontal cortical gray and white matter was extracted from the brain of an affected member of the PPND kindred as described (13). Hyperphosphorylated tau proteins in the insoluble fractions were dephosphorylated with alkaline phosphatase according to a published protocol (14). As a control, native tau samples were incubated identically except for the addition of 200 mM sodium pyrophosphate to inhibit alkaline phosphatase. For Western blot analyses, nitrocellulose replicas were prepared from 10% SDS/PAGE gels containing normal (see below) and abnormal tau proteins, and these replicas were probed with several well characterized mAbs specific for defined tau epitopes as reported (13). Paired helical filament tau (PHFtau) was extracted from brains of patients with AD as described (13), and a mixture of recombinant human tau proteins (a gift from M. Goedert, MRC Laboratory of Molecular Biology, Cambridge, England) containing all six isoforms of brain tau also was used as control samples.
RESULTS
Detection of Mutations in the Tau Gene.
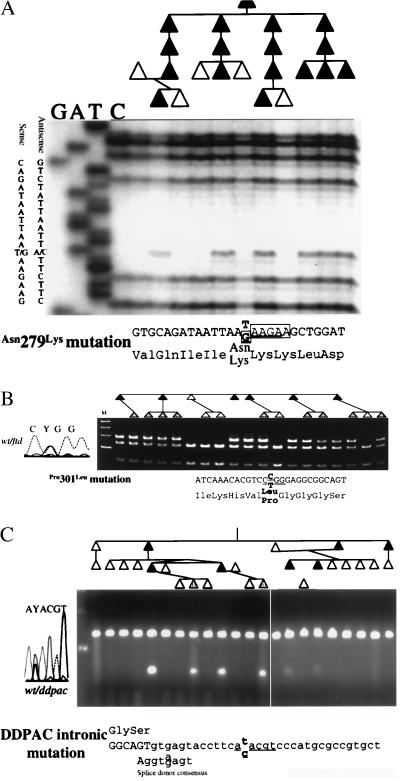
The tau gene was screened for mutations by DNA sequence analysis. Genomic DNA sequencing spanned all intron–exon boundaries for all tau exons, including those not normally found in human adult brain (exons 4a, 6, and 8) for subjects in PPND and DDPAC families. Genomic DNA from affected subjects in 65 non-AD kindreds were sequenced at least partially. Because FTDP-17 is autosomal dominant, affected subjects should be heterozygous for the pathogenic mutation. The PPND subject was heterozygous at 11 sites. Ten of these sites were variable in normal controls and were thus polymorphisms. The exon 10 variant, a T to G transversion, was not detected in 95 Caucasians and encoded a predicted change of an asparagine to a lysine at amino acid 279 in the second MT-binding domain. The 279Lys allele cosegregated with the disease in all affected subjects tested (n = 17, Figs. 1A and 2).
Figure 1.
Mutation analysis in PPND and other FTDP-17 syndromes. Pedigree and mutation analyses for three kindreds are shown. Direct DNA sequence (A) or restriction endonuclease cleavage analyses (B and C) are shown for three mutations. A shows the analysis for the PPND kindred with a Asn279Lys mutation. B shows the Pro301Leu mutation segregating in the Montreal family. C shows the segregation of the intronic mutation in the DDPAC family. The affected DNA sequence is shown below each panel. The underlined sequence is the recognition sequence for a restriction endonuclease. The potential exon-splicing-enhancer sequence is enclosed in boxes in A. Sexes have been hidden, and birth orders have been modified to maintain patient confidentiality. Individuals with shaded symbols are younger than the average age of onset in Table 1.
Figure 2.
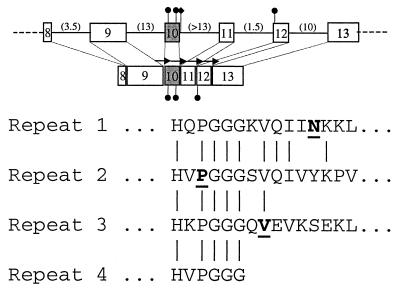
Tau mutations in PPND and other FTDP-17 syndromes. Organization of the carboxy-terminus of the tau gene is shown. Exons are shown as boxes. The numbers in parentheses are the size of the intron, in kilobases. Exon 10, which is spliced alternatively to produce a transcript for 3Rtau and 4Rtau, is shaded. Black circles indicate the locations of the four known missense mutations. The intronic DDPAC mutation is indicated by a black diamond. The repeated microtubule binding domain’s amino acid sequence is shown below. Mutated amino acids are indicated in bold. Arrows in the diagram represent the repeats.
An additional missense mutation was identified by DNA sequence analysis of two familial FTD kindreds (Montreal family and Seattle D Kindred). In exon 10, a C to T transition resulted in a predicted amino acid change of a proline to a leucine at the amino acid position in the second MT-binding domain and was conserved in all four MT domains in tau (Figs. 1A and 2). This 301Leu allele was not observed in 95 Caucasian controls. A panel of 111 unrelated affected subjects from familial dementia kindreds was screened for this mutation and for two that yielded additional 301Leu carriers (Seattle F and Oregon EL kindreds). In both cases, all affected family members available for sampling had the 301Leu mutation. The families with 301Leu are all of French Canadian ancestry. Genealogic studies and haplotype comparisons are underway to determine whether these families have inherited 301Leu from a common ancestor.
In the DDPAC kindred, no tau coding sequence mutations were identified. Heterozygous sites were detected in exons 4A (3 sites), 6 (2 sites), 7, 8, and 9 (4 sites), but all were polymorphisms (7). In noncoding regions, 12 heterozygous sites were detected, of which 11 were deemed polymorphisms based on their common occurrence among controls (information on polymorphic sites is available from the authors). However, a C to T transition that segregates with the disease and is not observed in 95 unrelated Caucasian controls was identified in intron 10, 14 bp after the 3′ end of exon 10 (Fig. 1C). The proximity of this site to the exon splice-donor site and its absence in normal controls suggest that this variant may be a pathogenic mutation that affects the regulation of exon 10 splicing.
Tau Isoforms in the Brains of FTDP-17 Families.
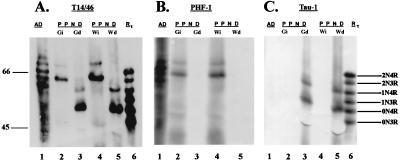
Filamentous and abnormal tau protein from affected subjects was studied to determine the functional consequences of the mutations observed. Previous studies have shown that tau isolated from the insoluble fraction (filamentous tau) of the brains from PPND subjects migrates as two bands with apparent Mr of 69 and 64 (4). To identify which tau isoforms are present in the insoluble fraction, tau was dephosphorylated, and the relative electrophoretic mobilities of the native and dephosphorylated tau proteins were compared with those of PHFtau isolated from brains of patients with AD and to all six isoforms of recombinant human tau (Fig. 3). Immunolabeling with a mixture of two phosphorylation-independent mAbs (T14 and T46) showed that the dephosphorylated tau protein from gray and white matter of the PPND brain exhibited accelerated electrophoretic mobility compared with native tau from the same brain (compare lanes 2 and 3 and lanes 4 and 5 in Fig. 3A). Furthermore, the dephosphorylated tau comigrated with the 4Rtau isoforms but not with the 3Rtau isoforms (compare lanes 5 and 6 in Fig. 3 A and C). The largest 4Rtau isoform (i.e., 2N4R) is not always detectable because of the low levels of this isoform even in the normal brain (see below) (14). Because previous studies have shown tau inclusions in neuronal perikarya in gray matter as well as glial cells in the white matter (4), we asked whether similar insoluble 4Rtau isoforms are recovered from gray and white matter. Fig. 3 shows that insoluble tau proteins recovered from both gray and white matter indeed are comprised exclusively of the 4Rtau isoforms (compare lanes Gi and Gd and lanes Wi and Wd with recombinant tau in lane 6 of Fig. 3 A and C). To confirm that native tau from PPND brain was dephosphorylated completely, identical nitrocellulose replicas were probed with PHF1, a phosphorylation-dependent mAb, and Tau1, a dephosphorylation-dependent mAb (13). As expected, native tau from PPND brain was immunolabeled with PHF1 (Fig. 3B) but not with Tau1 (Fig. 3C). In contrast, dephosphorylated tau from the same brain was stained by Tau1 but not by PHF1. Thus, in PPND brains, isoforms with four MT-binding repeat tau are aggregated as filamentous inclusions in gray and white matter.
Figure 3.
Western blots of native and dephosphorylated tau from a PPND brain. Insoluble fractions from gray and white matter were probed with the anti-tau mAb identified above each blot. Dephosphorylation reveals two predominant bands in tau from PPND brain that align with recombinant 4Rtau isoforms with an apparent molecular weight of 52 and 59. Molecular weight standards are shown in the left margin, and the positions of each of the nonphosphorylated recombinant human tau isoforms are labeled. Lane 1 shows PHFtau from AD brain. Lanes 2 and 3 show native and dephosphorylated tau, respectively, from gray matter of a PPND brain. Lanes 4 and 5 show native and dephosphorylated tau, respectively, from white matter of a PPND brain. Lane 6 shows all six recombinant tau isoforms (Rtau). Gi and Gd are native (i.e., insoluble) and dephosphorylated tau, respectively, from gray matter of a PPND brain. Wi and Wd are native and dephosphorylated tau, respectively, from white matter of a PPND brain. 0N3R, 1N3R, and 2N3R are three repeat tau isoforms with no vs. one or two amino terminal inserts, respectively. 0N4R, 1N4R, and 2N4R are four repeat tau isoforms with no vs. one or two amino terminal inserts, respectively.
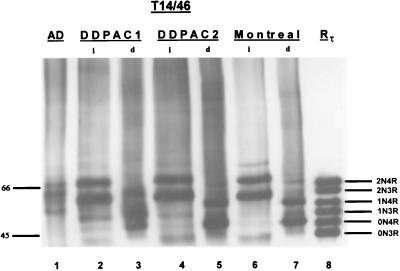
The tau isoforms present in the insoluble fraction also were examined in brain homogenates from the DDPAC and Montreal kindreds. Two major tau protein bands with apparent Mr of 69 and 64 and a minor band with Mr of ≈72 (Fig. 4, lanes 2, 4, and 6) were identified. This 72-kDa protein band probably is 4Rtau isoform with exons 2 and 3 added [i.e., 2N4Rtau (15)]. Dephosphorylation of insoluble tau from the DDPAC and the Montreal kindreds increased the electrophoretic mobility of the tau isoforms such that they comigrated with all three 4Rtau isoforms (compare lanes 2 and 3, 4 and 5, and 6 and 7 in Fig. 4). In one of the DDPAC cases (Fig. 4, lane 3) there appears to have been incomplete dephosphorylation. Thus, our data support the conclusion that all three mutations reported here resulted in the formation of aggregates of 4Rtau as insoluble aggregates.
Figure 4.
Western blots of native and dephosphorylated tau from the DDPAC and Montreal kindreds. Insoluble fractions obtained from the frontal cortex of two different brains of a DDPAC kindred and a brain from the Montreal kindred were probed with the two phosphorylation independent mAbs T14 and T46. Dephosphorylation reveals two major and one minor band that align with all recombinant 4Rtau isoforms (lanes 3, 5, and 7). The major bands align with 0N4Rtau and 1N4Rtau isoforms, and the minor band aligns with the 2N4Rtau isoform. Molecular weight standards are shown in the left margin, and the positions of each of the nonphosphorylated recombinant tau isoforms are labeled. Lane 1 shows PHFtau from the brain of a patient with AD. Lanes 2 and 4 show native tau, and lanes 3 and 5 show dephosphorylated tau from two different DDPAC brains. Lanes 6 and 7 show native and dephosphorylated tau from a brain of the Montreal kindred. Abbreviations are as in Fig. 3.
DISCUSSION
Genetic linkage studies localized the FTDP-17 gene to 17q21–22, the same region that contains the gene for tau. Because of these genetic data and because FTDP-17 brains have tau pathology, tau is a strong candidate gene for this disease. The first evidence that tau mutations can be pathogenic was reported by Poorkaj et al. (7), who found a tau missense mutation (Val337Met) that cosegregated with FTD in a large, well characterized family. We provide additional compelling evidence here that some FTDP-17 syndromes are caused by mutations in the tau gene. Specifically, we report two additional coding sequence missense mutations in exon 10 of the tau gene in affected members of PPND and four other FTDP-17 kindreds. The following evidence argues that the Asn279Lys and Pro301Leu variants described here and the Val337Met variant (7) are pathogenic mutations: (i) The mutations segregate with the disease in each family studied; (ii) the mutations are observed only in FTDP-17 kindreds (one family each for the Asn279Lys and the Val337Met mutations and in four families for the Pro301Leu mutation) and not in 95 controls; and (iii) the Pro301Leu change is in a functional domain (MT-binding repeat) and is conserved in all MT-binding repeats of humans and other species. The Val337Met mutation is also at a site in the inter-repeat sequences between the MT-binding repeats that is conserved in all three inter-repeats in human and mouse tau (7). Similarly, the Asn279Lys mutation is at an inter-repeat site conserved between human and mouse.
We also describe a strong association between an intronic nucleotide substitution 3′ to exon 10 in the tau gene and the disease state in the DDPAC kindred. The evidence that this mutation is pathogenic is as follows: (i) The mutation is found only in subjects affected with DDPAC and not in controls; and (ii) even though disease in this family clearly segregates with 17q21–22 markers, no other mutation was found in any coding exon. The location of this mutation is adjacent to the splice donor sequence for exon 10 (Fig. 2A), and thus it is difficult from the sequence alone to predict the functional consequences of the mutation. We propose that the mutation may affect the splicing to cause an increased production of 4Rtau. A similar argument was made by Conrad et al. (16), who reported an association between sporadic progressive supranuclear palsy and an intronic tau microsatellite polymorphism (17). In progressive supranuclear palsy, the forms of tau protein observed are predominantly 4Rtau, which correlates with an excess of tau mRNA containing exon 10 (18). Taken together, these findings suggest that genetic variation in potential regulatory sequences in tau introns or regions flanking the tau gene may play a role in the susceptibility of individuals to a variety of nonfamilial neurodegenerative syndromes.
Tau proteins in the human brain consist of six alternatively spliced isoforms with three or four imperfect tandem MT-binding repeats in the carboxy-terminal domain as well as one, two, or no amino-terminal inserts. Although the function of the amino-terminal inserts is unknown, the function of the carboxy-terminal MT-binding repeats is to bind and stabilize MTs. Thus, mutations in the MT binding repeats could reduce the binding of tau to MTs, resulting in the accumulation of excess tau as insoluble filamentous aggregates in the perikarya of neurons. The previously described Val337Met substitution is the result of a missense mutation in exon 12 that encodes the third MT binding repeat. If the Val337Met mutation disrupts binding of tau proteins to MTs, the binding of 3Rtau and 4Rtau to MTs should both be affected because the third MT binding repeat is present in all 3Rtau and 4Rtau isoforms. Previous studies have shown that the filamentous inclusions found in this kindred contain hyperphosphorylated tau isoforms (3Rtau and 4Rtau) that are biochemically and morphologically indistinguishable from PHFtau in AD (19). Because the PHFtau that forms PHFs in the neurofibrillary tangles of the AD brain contains all six tau isoforms (9), we infer that the Val337Met mutation alters the MT binding ability of all six tau isoforms such that all of the tau isoforms accumulate as insoluble PHFs in the brains of these patients.
Unlike the Val337Met mutation, all of the mutations found in the kindreds described here are either missense mutations in exon 10 or an intronic mutation near the 3′ splice site adjacent to exon 10. Because exon 10 encodes the second MT-binding repeat, which is found only in 4Rtau, our biochemical data demonstrating that 4Rtau isoforms accumulate as insoluble aggregates in the brains of these patients confirms that only those tau isoforms containing the MT-binding repeat encoded by exon 10 (i.e., 4Rtau) are affected. The mechanism whereby each of the mutations causes the formation of aggregates of 4Rtau may be quite different. For example, the Pro301 residue encoded by exon 10 is strongly conserved evolutionarily and is present in each of the four MT-binding repeats as part of a ProGlyGlyGly motif. Our finding that 4Rtau but not 3Rtau isoforms are recovered in the insoluble fraction is consistent with a reduced 4Rtau binding to MTs, leading to an increase in 4Rtau in the cytosol followed by the eventual aggregation of 4Rtau in cell bodies of neurons and glia. Although the Val337Met and Pro301Leu mutations could be pathogenic because of altered tau–microtubule binding properties, another hypothesis is that these mutations enhance the tau–tau interactions that occur when abnormal filaments are formed. Thus, the mutations would accelerate the formation of these filaments, which in turn could be toxic to specific cells.
The mechanism whereby the Asn279Lys mutation described here causes the aggregation of 4Rtau is less clear. This substitution in the carboxy-terminal portion of the second tandem MT binding inter-repeat changes an evolutionarily conserved residue in exon 10. However, Asn279 is not a conserved residue in other tandem MT binding inter-repeats. The other amino acid residues in the same location as Asn279 in other inter-repeats have uncharged polar residues (tyrosine, serine, and threonine). The Asn279Lys mutation leads to three adjacent lysine residues. Because previous studies have shown the importance of inter-repeats in regulating the binding of tau to MTs (20), the Asn279Lys mutation could reduce the binding of 4Rtau but not the 3Rtau to MTs. Our biochemical data on insoluble tau obtained from the PPND kindred are consistent with this prediction. However, another explanation is that this mutation affects the regulation of alternative splicing of exon 10 by creating or strengthening an exon-splicing-enhancer sequence within exon 10. Exon-splicing-enhancers are polypurine rich and contain a common 6- to 9-bp repeat of the sequence GAR, where R is a purine (21–23). The Asn279Lys mutation changes the normal sequence of TAAGAA to GAAGAA, a potential GAR exon-splicing-enhancer motif. The result in PPND subjects may be increased inclusion of exon 10 in tau mRNA and thus overproduction of 4Rtau proteins. This hypothesis is also consistent with our biochemical data that the insoluble tau aggregates from PPND brain predominantly contain 4Rtau.
Finally, the identification of an intronic mutation adjacent to the splice donor consensus sequence of exon 10 in the tau gene suggests a possible defect in the regulation of exon 10 splicing. Because this C to T intronic mutation resides within the “stem” of a short potential stem–loop structure (12-base stem, 6-base loop) that spanned the splice site of exon 10, it could lead to destabilization of the stem–loop structure, resulting in increased production of mRNA with exon 10. This in turn will lead to an increased production of 4Rtau.
Our biochemical data demonstrate that exon 10 mutations in the tau gene result in the accumulation of the 4Rtau isoforms as filamentous inclusions in the brains of some affected FTDP-17 kindreds. We therefore propose that the mutations in the tau gene of such individuals play a mechanistic role in the onset and progression of this FTDP-17 syndrome by inducing the aggregation of tau isoforms with four MT binding repeats into the hallmark lesions of the FTDP-17 brain. Additional studies are needed to test this hypothesis. The identification of three missense mutations in the tandem MT binding repeats and an intronic mutation directly implicate tau in mechanisms leading to hereditary FTDP-17 neurodegenerative syndromes. Finally, although pathogenic mutations in the tau gene have not been reported in familial AD, we speculate that epigenetic alterations in the biophysical properties of wild-type tau also might play a mechanistic role in the onset and progression of sporadic forms of AD and related neurodegenerative diseases.
Acknowledgments
The authors thank the patients that participated in this research. This work was supported in part from National Institute on Aging Grants AF1176-03 (to G.D.S.), AG-10124 (to J.Q.T.), AG-10210 (to V.M.Y.L.), AG-09215 (to J.Q.T.), AG14382 (to V.M.Y.L. and G.D.S.), AG-14449 (to J.Q.T.), and AG-08017 (to H.P.), Veterans Affairs Administration Merit Award 0011 (to G.D.S.), a grant from the University of California at Los Angeles Alzheimer’s disease center (to D.H.G.), National Institute on Neurological Disorders and Stroke Grants NS31212-05 (to K.C.W.) and NS36733-01A1 (to K.C.W.), pilot research support from the University of California at Davis Alzheimer’s Center (to K.C.W.), and intramural support from the Ernest Gallo Clinic and Research Foundation (to K.C.W.). Support for identification of the A and B families is from the National Institute on Aging Alzheimer’s Disease Research Center (Grant AG05136).
ABBREVIATIONS
- FTD
frontotemporal dementia
- FTDP
FTD and parkinsonism
- DDPAC
disinhibition–dementia–parkinsonism–amyotrophy complex
- PPND
pallido-ponto-nigral degeneration
- MT
microtubule
- AD
Alzheimer’s disease
- PHF
paired helical filament
Note
Since the initial submission of this manuscript, two papers have identified additional mutations in the tau gene that produce familial dementia (24, 25). Hutton et al. (25) identify additional families with the Pro301Leu mutations. Both papers identify mutations in the 3′ prime region of exon 10 that are posited to affect RNA splicing.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1. Foster N L, Wilhelmsen K C, Sima A A F, Jones M Z, D’Amato C, Gilman S. Ann Neurol. 1997;41:706–715. doi: 10.1002/ana.410410606. [DOI] [PubMed] [Google Scholar]
- 2.Wilhelmsen K C, Lynch T, Nygaard T G. Am J Hum Genet. 1994;55:1159–1165. [PMC free article] [PubMed] [Google Scholar]
- 3.Wszolek Z K, Pfeiffer R F, Bhatt M H, Schelper R L, Cordes M, Snow B J, Rodnitzky R L, Wolters E C, Arwert F, Calne D B. Ann Neurol. 1992;32:312–320. doi: 10.1002/ana.410320303. [DOI] [PubMed] [Google Scholar]
- 4.Reed L A, Schmidt M L, Wszolek Z K, Balin B J, Soontornniyomkij V, Lee V M Y, Trojanowski J Q. J Neuropathol Exp Neurol. 1998;57:588–601. doi: 10.1097/00005072-199806000-00006. [DOI] [PubMed] [Google Scholar]
- 5.Froelich S, Basun H, Forsell C, Lilius L, Axelman K, Andreadis A, Lannfelt L. Am J Med Genet. 1997;74:380–385. doi: 10.1002/(sici)1096-8628(19970725)74:4<380::aid-ajmg8>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 6.Baker M, Kwok J B, Kucera S, Crook R, Farrer M, Houlden H, Isaacs A, Lincoln S, Onstead L, Hardy J, et al. Ann Neurol. 1997;42:794–798. doi: 10.1002/ana.410420516. [DOI] [PubMed] [Google Scholar]
- 7.Poorkaj P, Bird T, Wijsman E, Nemens E, Garruto R M, Anderson L, Andreadis A, Wiederholt W C, Raskind M, Schellenberg G D. Ann Neurol. 1998;43:815–825. doi: 10.1002/ana.410430617. [DOI] [PubMed] [Google Scholar]
- 8.Goedert M, Spillantini M G, Jakes R, Rutherford D, Crowther R A. Neuron. 1989;3:519–526. doi: 10.1016/0896-6273(89)90210-9. [DOI] [PubMed] [Google Scholar]
- 9.Goedert M, Spillantini M G, Cairns N J, Crowther R A. Neuron. 1992;8:159–168. doi: 10.1016/0896-6273(92)90117-v. [DOI] [PubMed] [Google Scholar]
- 10.Wszolek Z K, Pfeiffer R F. In: Parkinsonian Syndromes. Stern M, Koller W C, editors. New York: Marcel Dekker; 1993. pp. 297–312. [Google Scholar]
- 11.Denson M A, Wszolek Z K. Parkinsonism and Related Disorders. 1995;1:35–46. doi: 10.1016/1353-8020(95)00010-4. [DOI] [PubMed] [Google Scholar]
- 12.Wszolek Z K, Lynch T, Wilhelmsen K C. Parkinsonism and Related Disorders. 1997;3:67–76. doi: 10.1016/s1353-8020(97)00006-0. [DOI] [PubMed] [Google Scholar]
- 13.Bramblett G T, Goedert M, Jakes R, Merrick S E, Trojanowski J Q, Lee V M Y. Neuron. 1993;10:1089–1099. doi: 10.1016/0896-6273(93)90057-x. [DOI] [PubMed] [Google Scholar]
- 14.Goedert M, Spillantini M G, Cairns N J, Crowther R A. Neuron. 1992;8:159–168. doi: 10.1016/0896-6273(92)90117-v. [DOI] [PubMed] [Google Scholar]
- 15.Goedert M, Spillantini M G, Jakes R, Rutherford D, Crowther R A. Neuron. 1989;3:519–526. doi: 10.1016/0896-6273(89)90210-9. [DOI] [PubMed] [Google Scholar]
- 16.Conrad C, Andreadis A, Trojanowski J Q, Dickson D W, Kang D, Chen X, Wiederholt W, Hansen L, Masliah E, Thal L J, et al. Ann Neurol. 1997;41:277–281. doi: 10.1002/ana.410410222. [DOI] [PubMed] [Google Scholar]
- 17.Higgin J J, Litvan I, Pho L T, Li W, Nee L E. Neurology. 1998;50:270–273. doi: 10.1212/wnl.50.1.270. [DOI] [PubMed] [Google Scholar]
- 18.Chambers C B, Lee J M, Muma N A. J Neuropathol Exp Neurol. 1998;57:474. [Google Scholar]
- 19.Spillantini M G, Crowther R A, Goedert M. Acta Neuropathol. 1996;92:42–48. doi: 10.1007/s004010050487. [DOI] [PubMed] [Google Scholar]
- 20.Goode B L, Feinstein S C. J Cell Biol. 1994;124:769–782. doi: 10.1083/jcb.124.5.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu R, Teng J, Cooper T A. Mol Cell Biol. 1993;13:3660–3674. doi: 10.1128/mcb.13.6.3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Watakabe A, Tanaka K, Shimura Y. Genes Dev. 1993;7:407–418. doi: 10.1101/gad.7.3.407. [DOI] [PubMed] [Google Scholar]
- 23.Lavigueur A, La Branche H, Kornblihtt A R, Chabot B. Genes Dev. 1993;7:2405–2417. doi: 10.1101/gad.7.12a.2405. [DOI] [PubMed] [Google Scholar]
- 24.Spillantini M G, Murrell J R, Goedert M, Farlow M R, Klug A, Ghetti B. Proc Natl Acad Sci USA. 1998;95:7737–7741. doi: 10.1073/pnas.95.13.7737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hutton M, Lendon C L, Rizzu P, Baker M, Froelich S, Houlden H, Pickering-Brown S, Chakrarerty S, Isaacs A, Grover A, et al. Nature (London) 1998;393:702–705. doi: 10.1038/31508. [DOI] [PubMed] [Google Scholar]