Abstract
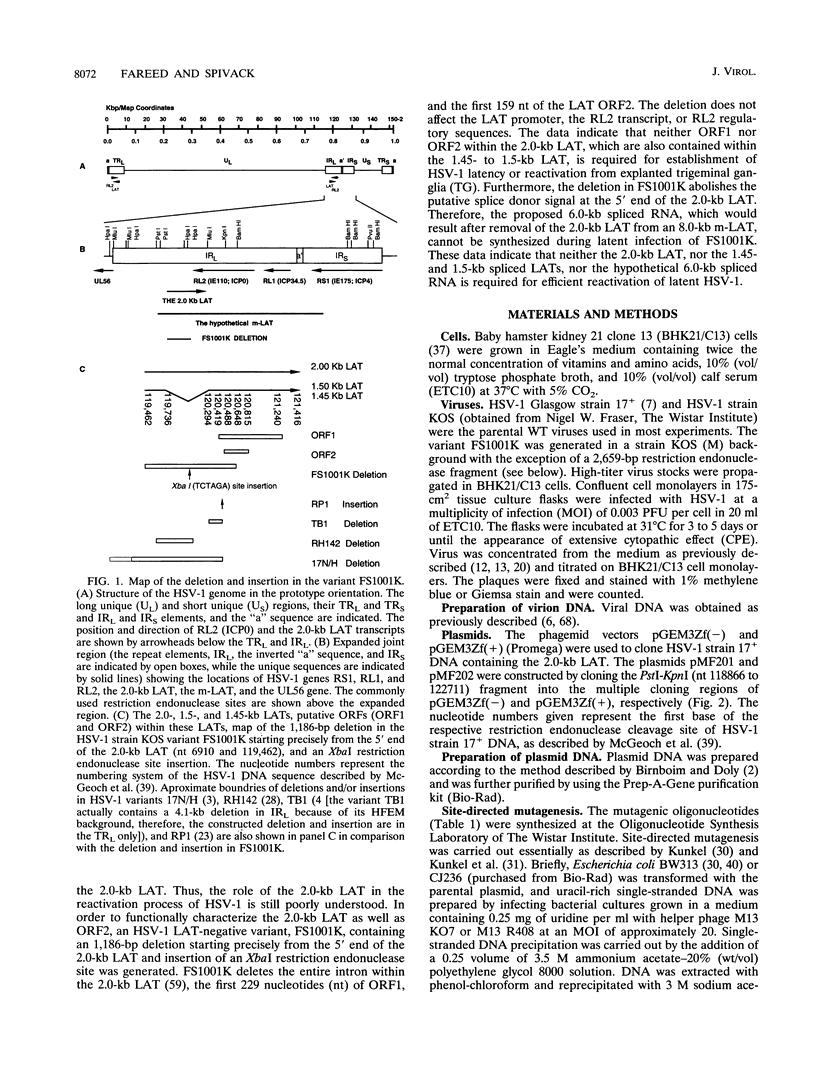
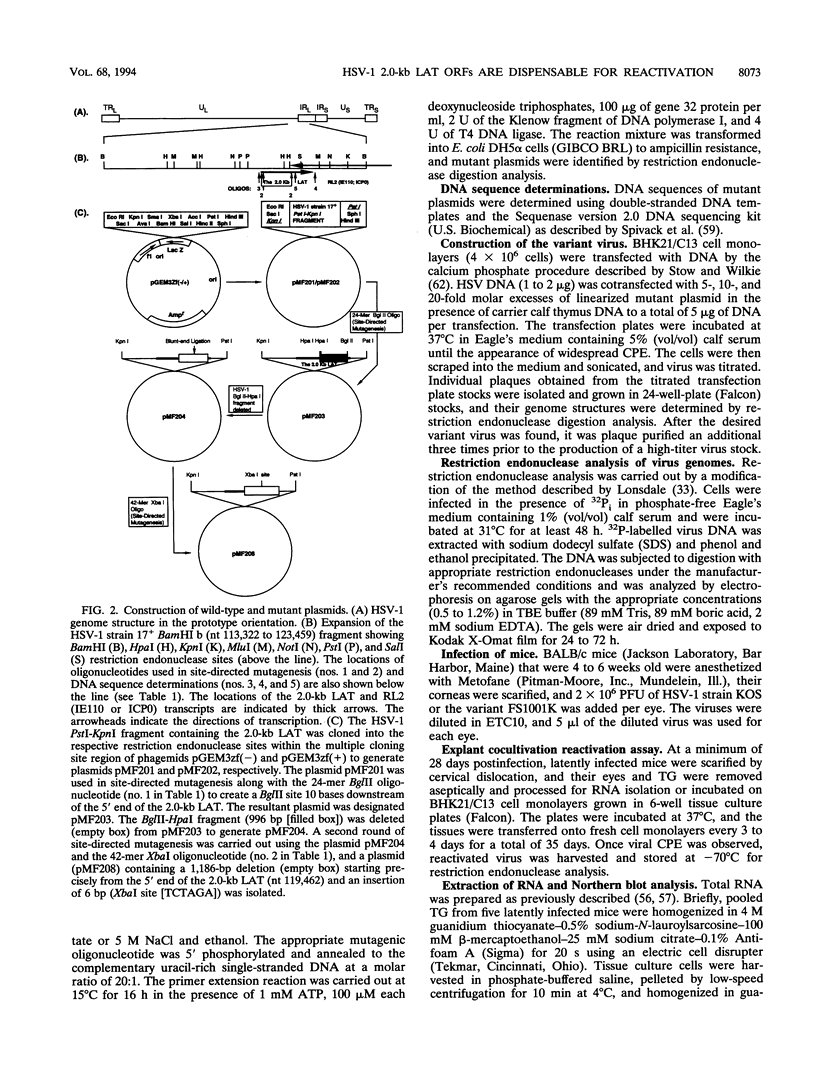
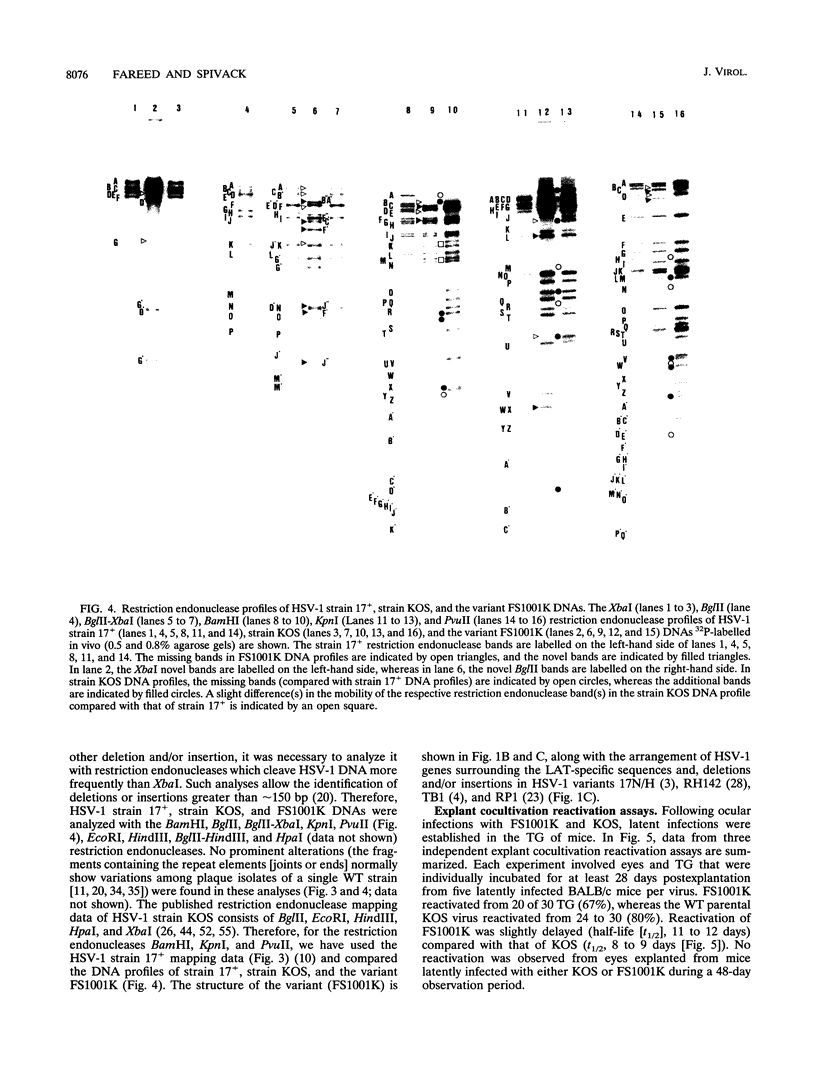
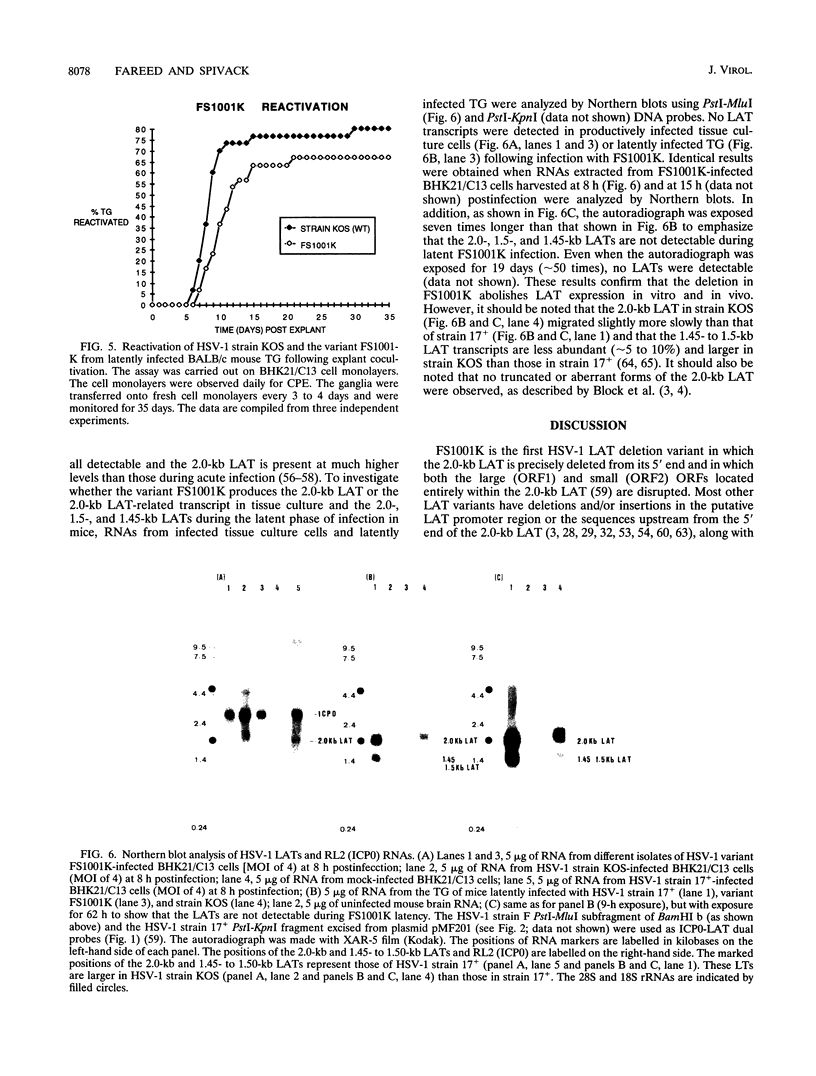
The herpes simplex virus type 1 (HSV-1) latency-associated transcripts (LATs) are dispensable for establishment and maintenance of latent infection. However, the LATs have been implicated in reactivation of the virus from its latent state. Since the reported LAT deletion and/or insertion variants that are reactivation impaired contain deletions in the putative LAT promoter, it is not known which LAT sequences are involved in reactivation. To examine the role of the 2.0-kb LAT in the process of reactivation and the functional importance of the putative open reading frames (ORF1 and ORF2) contained within the 2.0-kb LAT, we have constructed an HSV-1 variant that contains a precise deletion and insertion within the LAT-specific DNA sequences using site-directed mutagenesis. The HSV-1 variant FS1001K contains an 1,186-bp deletion starting precisely from the 5' end of the 2.0-kb LAT and, for identification, a XbaI restriction endonuclease site insertion. The FS1001K genome contains no other deletions and/or insertions as analyzed by a variety of restriction endonucleases. The deletion in FS1001K removes the entire 556-bp intron within the 2.0-kb LAT, the first 229 nucleotides of ORF1, and the first 159 nucleotides of ORF2 without having an affect on the RL2 (ICP0) gene. Explant cocultivation reactivation assays indicated that this deletion had a minimal effect on reactivation of the variant FS1001K compared with the parental wild-type virus using a mouse eye model. As expected, Northern (RNA) blot analyses have shown that the variant virus (FS1001K) does not produce the 2.0-kb LAT or the 1.45- to 1.5-kb LAT either in vitro or in vivo; however, FS1001K produces an intact RL2 transcript in tissue culture. These data suggest that the 2.0-kb LAT putative ORF1 and ORF2 (or the first 1,186 bp of the 2.0-kb LAT) are dispensable for explant reactivation of latent HSV-1.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baines J. D., Roizman B. The open reading frames UL3, UL4, UL10, and UL16 are dispensable for the replication of herpes simplex virus 1 in cell culture. J Virol. 1991 Feb;65(2):938–944. doi: 10.1128/jvi.65.2.938-944.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block T. M., Deshmane S., Masonis J., Maggioncalda J., Valyi-Nagi T., Fraser N. W. An HSV LAT null mutant reactivates slowly from latent infection and makes small plaques on CV-1 monolayers. Virology. 1993 Feb;192(2):618–630. doi: 10.1006/viro.1993.1078. [DOI] [PubMed] [Google Scholar]
- Block T. M., Spivack J. G., Steiner I., Deshmane S., McIntosh M. T., Lirette R. P., Fraser N. W. A herpes simplex virus type 1 latency-associated transcript mutant reactivates with normal kinetics from latent infection. J Virol. 1990 Jul;64(7):3417–3426. doi: 10.1128/jvi.64.7.3417-3426.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom D. C., Devi-Rao G. B., Hill J. M., Stevens J. G., Wagner E. K. Molecular analysis of herpes simplex virus type 1 during epinephrine-induced reactivation of latently infected rabbits in vivo. J Virol. 1994 Mar;68(3):1283–1292. doi: 10.1128/jvi.68.3.1283-1292.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown S. M., Harland J., Subak-Sharpe J. H. Isolation of restriction endonuclease site deletion mutants of herpes simplex virus. J Gen Virol. 1984 Jun;65(Pt 6):1053–1068. doi: 10.1099/0022-1317-65-6-1053. [DOI] [PubMed] [Google Scholar]
- Brown S. M., Ritchie D. A., Subak-Sharpe J. H. Genetic studies with herpes simplex virus type 1. The isolation of temperature-sensitive mutants, their arrangement into complementation groups and recombination analysis leading to a linkage map. J Gen Virol. 1973 Mar;18(3):329–346. doi: 10.1099/0022-1317-18-3-329. [DOI] [PubMed] [Google Scholar]
- Chou J., Kern E. R., Whitley R. J., Roizman B. Mapping of herpes simplex virus-1 neurovirulence to gamma 134.5, a gene nonessential for growth in culture. Science. 1990 Nov 30;250(4985):1262–1266. doi: 10.1126/science.2173860. [DOI] [PubMed] [Google Scholar]
- Copertino D. W., Hallick R. B. Group II twintron: an intron within an intron in a chloroplast cytochrome b-559 gene. EMBO J. 1991 Feb;10(2):433–442. doi: 10.1002/j.1460-2075.1991.tb07965.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison A. J., Wilkie N. M. Nucleotide sequences of the joint between the L and S segments of herpes simplex virus types 1 and 2. J Gen Virol. 1981 Aug;55(Pt 2):315–331. doi: 10.1099/0022-1317-55-2-315. [DOI] [PubMed] [Google Scholar]
- Deatly A. M., Spivack J. G., Lavi E., Fraser N. W. RNA from an immediate early region of the type 1 herpes simplex virus genome is present in the trigeminal ganglia of latently infected mice. Proc Natl Acad Sci U S A. 1987 May;84(10):3204–3208. doi: 10.1073/pnas.84.10.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deatly A. M., Spivack J. G., Lavi E., O'Boyle D. R., 2nd, Fraser N. W. Latent herpes simplex virus type 1 transcripts in peripheral and central nervous system tissues of mice map to similar regions of the viral genome. J Virol. 1988 Mar;62(3):749–756. doi: 10.1128/jvi.62.3.749-756.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshmane S. L., Fraser N. W. During latency, herpes simplex virus type 1 DNA is associated with nucleosomes in a chromatin structure. J Virol. 1989 Feb;63(2):943–947. doi: 10.1128/jvi.63.2.943-947.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshmane S. L., Nicosia M., Valyi-Nagy T., Feldman L. T., Dillner A., Fraser N. W. An HSV-1 mutant lacking the LAT TATA element reactivates normally in explant cocultivation. Virology. 1993 Oct;196(2):868–872. doi: 10.1006/viro.1993.1548. [DOI] [PubMed] [Google Scholar]
- Devi-Rao G. B., Bloom D. C., Stevens J. G., Wagner E. K. Herpes simplex virus type 1 DNA replication and gene expression during explant-induced reactivation of latently infected murine sensory ganglia. J Virol. 1994 Mar;68(3):1271–1282. doi: 10.1128/jvi.68.3.1271-1282.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson A. T., Sederati F., Devi-Rao G., Flanagan W. M., Farrell M. J., Stevens J. G., Wagner E. K., Feldman L. T. Identification of the latency-associated transcript promoter by expression of rabbit beta-globin mRNA in mouse sensory nerve ganglia latently infected with a recombinant herpes simplex virus. J Virol. 1989 Sep;63(9):3844–3851. doi: 10.1128/jvi.63.9.3844-3851.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerig C., Pizer L. I., Wilcox C. L. An antigen encoded by the latency-associated transcript in neuronal cell cultures latently infected with herpes simplex virus type 1. J Virol. 1991 May;65(5):2724–2727. doi: 10.1128/jvi.65.5.2724-2727.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efstathiou S., Minson A. C., Field H. J., Anderson J. R., Wildy P. Detection of herpes simplex virus-specific DNA sequences in latently infected mice and in humans. J Virol. 1986 Feb;57(2):446–455. doi: 10.1128/jvi.57.2.446-455.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell M. J., Dobson A. T., Feldman L. T. Herpes simplex virus latency-associated transcript is a stable intron. Proc Natl Acad Sci U S A. 1991 Feb 1;88(3):790–794. doi: 10.1073/pnas.88.3.790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell M. J., Hill J. M., Margolis T. P., Stevens J. G., Wagner E. K., Feldman L. T. The herpes simplex virus type 1 reactivation function lies outside the latency-associated transcript open reading frame ORF-2. J Virol. 1993 Jun;67(6):3653–3655. doi: 10.1128/jvi.67.6.3653-3655.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser N. W., Block T. M., Spivack J. G. The latency-associated transcripts of herpes simplex virus: RNA in search of function. Virology. 1992 Nov;191(1):1–8. doi: 10.1016/0042-6822(92)90160-q. [DOI] [PubMed] [Google Scholar]
- Goins W. F., Sternberg L. R., Croen K. D., Krause P. R., Hendricks R. L., Fink D. J., Straus S. E., Levine M., Glorioso J. C. A novel latency-active promoter is contained within the herpes simplex virus type 1 UL flanking repeats. J Virol. 1994 Apr;68(4):2239–2252. doi: 10.1128/jvi.68.4.2239-2252.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldin A. L., Sandri-Goldin R. M., Levine M., Glorioso J. C. Cloning of herpes simplex virus type 1 sequences representing the whole genome. J Virol. 1981 Apr;38(1):50–58. doi: 10.1128/jvi.38.1.50-58.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill J. M., Sedarati F., Javier R. T., Wagner E. K., Stevens J. G. Herpes simplex virus latent phase transcription facilitates in vivo reactivation. Virology. 1990 Jan;174(1):117–125. doi: 10.1016/0042-6822(90)90060-5. [DOI] [PubMed] [Google Scholar]
- Ho D. Y., Mocarski E. S. Herpes simplex virus latent RNA (LAT) is not required for latent infection in the mouse. Proc Natl Acad Sci U S A. 1989 Oct;86(19):7596–7600. doi: 10.1073/pnas.86.19.7596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javier R. T., Stevens J. G., Dissette V. B., Wagner E. K. A herpes simplex virus transcript abundant in latently infected neurons is dispensable for establishment of the latent state. Virology. 1988 Sep;166(1):254–257. doi: 10.1016/0042-6822(88)90169-9. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Leib D. A., Bogard C. L., Kosz-Vnenchak M., Hicks K. A., Coen D. M., Knipe D. M., Schaffer P. A. A deletion mutant of the latency-associated transcript of herpes simplex virus type 1 reactivates from the latent state with reduced frequency. J Virol. 1989 Jul;63(7):2893–2900. doi: 10.1128/jvi.63.7.2893-2900.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonsdale D. M. A rapid technique for distinguishing herpes-simplex virus type 1 from type 2 by restriction-enzyme technology. Lancet. 1979 Apr 21;1(8121):849–852. doi: 10.1016/s0140-6736(79)91265-0. [DOI] [PubMed] [Google Scholar]
- Lonsdale D. M., Brown S. M., Lang J., Subak-Sharpe J. H., Koprowski H., Warren K. G. Variations in herpes simplex virus isolated from human ganglia and a study of clonal variation in HSV-1. Ann N Y Acad Sci. 1980;354:291–308. doi: 10.1111/j.1749-6632.1980.tb27973.x. [DOI] [PubMed] [Google Scholar]
- MACPHERSON I., STOKER M. Polyoma transformation of hamster cell clones--an investigation of genetic factors affecting cell competence. Virology. 1962 Feb;16:147–151. doi: 10.1016/0042-6822(62)90290-8. [DOI] [PubMed] [Google Scholar]
- MacLean A. R., Brown S. M. A herpes simplex virus type 1 variant which fails to synthesize immediate early polypeptide VmwIE63. J Gen Virol. 1987 May;68(Pt 5):1339–1350. doi: 10.1099/0022-1317-68-5-1339. [DOI] [PubMed] [Google Scholar]
- MacLean A. R., ul-Fareed M., Robertson L., Harland J., Brown S. M. Herpes simplex virus type 1 deletion variants 1714 and 1716 pinpoint neurovirulence-related sequences in Glasgow strain 17+ between immediate early gene 1 and the 'a' sequence. J Gen Virol. 1991 Mar;72(Pt 3):631–639. doi: 10.1099/0022-1317-72-3-631. [DOI] [PubMed] [Google Scholar]
- Margolis T. P., Bloom D. C., Dobson A. T., Feldman L. T., Stevens J. G. Decreased reporter gene expression during latent infection with HSV LAT promoter constructs. Virology. 1993 Dec;197(2):585–592. doi: 10.1006/viro.1993.1632. [DOI] [PubMed] [Google Scholar]
- McGeoch D. J., Dalrymple M. A., Davison A. J., Dolan A., Frame M. C., McNab D., Perry L. J., Scott J. E., Taylor P. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J Gen Virol. 1988 Jul;69(Pt 7):1531–1574. doi: 10.1099/0022-1317-69-7-1531. [DOI] [PubMed] [Google Scholar]
- Mead D. A., Szczesna-Skorupa E., Kemper B. Single-stranded DNA 'blue' T7 promoter plasmids: a versatile tandem promoter system for cloning and protein engineering. Protein Eng. 1986 Oct-Nov;1(1):67–74. doi: 10.1093/protein/1.1.67. [DOI] [PubMed] [Google Scholar]
- Mellerick D. M., Fraser N. W. Physical state of the latent herpes simplex virus genome in a mouse model system: evidence suggesting an episomal state. Virology. 1987 Jun;158(2):265–275. doi: 10.1016/0042-6822(87)90198-x. [DOI] [PubMed] [Google Scholar]
- Mitchell W. J., Lirette R. P., Fraser N. W. Mapping of low abundance latency-associated RNA in the trigeminal ganglia of mice latently infected with herpes simplex virus type 1. J Gen Virol. 1990 Jan;71(Pt 1):125–132. doi: 10.1099/0022-1317-71-1-125. [DOI] [PubMed] [Google Scholar]
- Mitchell W. J., Steiner I., Brown S. M., MacLean A. R., Subak-Sharpe J. H., Fraser N. W. A herpes simplex virus type 1 variant, deleted in the promoter region of the latency-associated transcripts, does not produce any detectable minor RNA species during latency in the mouse trigeminal ganglion. J Gen Virol. 1990 Apr;71(Pt 4):953–957. doi: 10.1099/0022-1317-71-4-953. [DOI] [PubMed] [Google Scholar]
- Morse L. S., Buchman T. G., Roizman B., Schaffer P. A. Anatomy of herpes simplex virus DNA. IX. Apparent exclusion of some parental DNA arrangements in the generation of intertypic (HSV-1 X HSV-2) recombinants. J Virol. 1977 Oct;24(1):231–248. doi: 10.1128/jvi.24.1.231-248.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullaney J., Moss H. W., McGeoch D. J. Gene UL2 of herpes simplex virus type 1 encodes a uracil-DNA glycosylase. J Gen Virol. 1989 Feb;70(Pt 2):449–454. doi: 10.1099/0022-1317-70-2-449. [DOI] [PubMed] [Google Scholar]
- Nicosia M., Deshmane S. L., Zabolotny J. M., Valyi-Nagy T., Fraser N. W. Herpes simplex virus type 1 latency-associated transcript (LAT) promoter deletion mutants can express a 2-kilobase transcript mapping to the LAT region. J Virol. 1993 Dec;67(12):7276–7283. doi: 10.1128/jvi.67.12.7276-7283.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post L. E., Roizman B. A generalized technique for deletion of specific genes in large genomes: alpha gene 22 of herpes simplex virus 1 is not essential for growth. Cell. 1981 Jul;25(1):227–232. doi: 10.1016/0092-8674(81)90247-6. [DOI] [PubMed] [Google Scholar]
- Puga A., Rosenthal J. D., Openshaw H., Notkins A. L. Herpes simplex virus DNA and mRNA sequences in acutely and chronically infected trigeminal ganglia of mice. Virology. 1978 Aug;89(1):102–111. doi: 10.1016/0042-6822(78)90044-2. [DOI] [PubMed] [Google Scholar]
- Rock D. L., Fraser N. W. Detection of HSV-1 genome in central nervous system of latently infected mice. Nature. 1983 Apr 7;302(5908):523–525. doi: 10.1038/302523a0. [DOI] [PubMed] [Google Scholar]
- Rock D. L., Nesburn A. B., Ghiasi H., Ong J., Lewis T. L., Lokensgard J. R., Wechsler S. L. Detection of latency-related viral RNAs in trigeminal ganglia of rabbits latently infected with herpes simplex virus type 1. J Virol. 1987 Dec;61(12):3820–3826. doi: 10.1128/jvi.61.12.3820-3826.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandri-Goldin R. M., Levine M., Glorioso J. C. Method for induction of mutations in physically defined regions of the herpes simplex virus genome. J Virol. 1981 Apr;38(1):41–49. doi: 10.1128/jvi.38.1.41-49.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawtell N. M., Thompson R. L. Herpes simplex virus type 1 latency-associated transcription unit promotes anatomical site-dependent establishment and reactivation from latency. J Virol. 1992 Apr;66(4):2157–2169. doi: 10.1128/jvi.66.4.2157-2169.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedarati F., Izumi K. M., Wagner E. K., Stevens J. G. Herpes simplex virus type 1 latency-associated transcription plays no role in establishment or maintenance of a latent infection in murine sensory neurons. J Virol. 1989 Oct;63(10):4455–4458. doi: 10.1128/jvi.63.10.4455-4458.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skare J., Summers W. C. Structure and function of herpesvirus genomes. II. EcoRl, Sbal, and HindIII endonuclease cleavage sites on herpes simplex virus. Virology. 1977 Feb;76(2):581–595. doi: 10.1016/0042-6822(77)90240-9. [DOI] [PubMed] [Google Scholar]
- Spivack J. G., Fraser N. W. Detection of herpes simplex virus type 1 transcripts during latent infection in mice. J Virol. 1987 Dec;61(12):3841–3847. doi: 10.1128/jvi.61.12.3841-3847.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spivack J. G., Fraser N. W. Expression of herpes simplex virus type 1 (HSV-1) latency-associated transcripts and transcripts affected by the deletion in avirulent mutant HFEM: evidence for a new class of HSV-1 genes. J Virol. 1988 Sep;62(9):3281–3287. doi: 10.1128/jvi.62.9.3281-3287.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spivack J. G., Fraser N. W. Expression of herpes simplex virus type 1 latency-associated transcripts in the trigeminal ganglia of mice during acute infection and reactivation of latent infection. J Virol. 1988 May;62(5):1479–1485. doi: 10.1128/jvi.62.5.1479-1485.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spivack J. G., Woods G. M., Fraser N. W. Identification of a novel latency-specific splice donor signal within the herpes simplex virus type 1 2.0-kilobase latency-associated transcript (LAT): translation inhibition of LAT open reading frames by the intron within the 2.0-kilobase LAT. J Virol. 1991 Dec;65(12):6800–6810. doi: 10.1128/jvi.65.12.6800-6810.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner I., Spivack J. G., Lirette R. P., Brown S. M., MacLean A. R., Subak-Sharpe J. H., Fraser N. W. Herpes simplex virus type 1 latency-associated transcripts are evidently not essential for latent infection. EMBO J. 1989 Feb;8(2):505–511. doi: 10.1002/j.1460-2075.1989.tb03404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens J. G., Wagner E. K., Devi-Rao G. B., Cook M. L., Feldman L. T. RNA complementary to a herpesvirus alpha gene mRNA is prominent in latently infected neurons. Science. 1987 Feb 27;235(4792):1056–1059. doi: 10.1126/science.2434993. [DOI] [PubMed] [Google Scholar]
- Stow N. D., Wilkie N. M. An improved technique for obtaining enhanced infectivity with herpes simplex virus type 1 DNA. J Gen Virol. 1976 Dec;33(3):447–458. doi: 10.1099/0022-1317-33-3-447. [DOI] [PubMed] [Google Scholar]
- Trousdale M. D., Steiner I., Spivack J. G., Deshmane S. L., Brown S. M., MacLean A. R., Subak-Sharpe J. H., Fraser N. W. In vivo and in vitro reactivation impairment of a herpes simplex virus type 1 latency-associated transcript variant in a rabbit eye model. J Virol. 1991 Dec;65(12):6989–6993. doi: 10.1128/jvi.65.12.6989-6993.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner E. K., Devi-Rao G., Feldman L. T., Dobson A. T., Zhang Y. F., Flanagan W. M., Stevens J. G. Physical characterization of the herpes simplex virus latency-associated transcript in neurons. J Virol. 1988 Apr;62(4):1194–1202. doi: 10.1128/jvi.62.4.1194-1202.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner E. K., Flanagan W. M., Devi-Rao G., Zhang Y. F., Hill J. M., Anderson K. P., Stevens J. G. The herpes simplex virus latency-associated transcript is spliced during the latent phase of infection. J Virol. 1988 Dec;62(12):4577–4585. doi: 10.1128/jvi.62.12.4577-4585.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wechsler S. L., Nesburn A. B., Watson R., Slanina S. M., Ghiasi H. Fine mapping of the latency-related gene of herpes simplex virus type 1: alternative splicing produces distinct latency-related RNAs containing open reading frames. J Virol. 1988 Nov;62(11):4051–4058. doi: 10.1128/jvi.62.11.4051-4058.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wechsler S. L., Nesburn A. B., Zwaagstra J., Ghiasi H. Sequence of the latency-related gene of herpes simplex virus type 1. Virology. 1989 Jan;168(1):168–172. doi: 10.1016/0042-6822(89)90416-9. [DOI] [PubMed] [Google Scholar]
- Wilkie N. M., Cortini R., Clements J. B. Structural studies and physical maps for the herpes simplex virus genome. J Antimicrob Chemother. 1977 Mar;3 (Suppl A):47–62. doi: 10.1093/jac/3.suppl_a.47. [DOI] [PubMed] [Google Scholar]
- Wilkie N. M. Physical maps for Herpes simplex virus type 1 DNA for restriction endonucleases Hind III, Hpa-1, and X. bad. J Virol. 1976 Oct;20(1):222–233. doi: 10.1128/jvi.20.1.222-233.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkie N. M. The synthesis and substructure of herpesvirus DNA: the distribution of alkali-labile single strand interruptions in HSV-1 DNA. J Gen Virol. 1973 Dec;21(3):453–467. doi: 10.1099/0022-1317-21-3-453. [DOI] [PubMed] [Google Scholar]
- Zwaagstra J. C., Ghiasi H., Slanina S. M., Nesburn A. B., Wheatley S. C., Lillycrop K., Wood J., Latchman D. S., Patel K., Wechsler S. L. Activity of herpes simplex virus type 1 latency-associated transcript (LAT) promoter in neuron-derived cells: evidence for neuron specificity and for a large LAT transcript. J Virol. 1990 Oct;64(10):5019–5028. doi: 10.1128/jvi.64.10.5019-5028.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwaagstra J., Ghiasi H., Nesburn A. B., Wechsler S. L. In vitro promoter activity associated with the latency-associated transcript gene of herpes simplex virus type 1. J Gen Virol. 1989 Aug;70(Pt 8):2163–2169. doi: 10.1099/0022-1317-70-8-2163. [DOI] [PubMed] [Google Scholar]





