Abstract
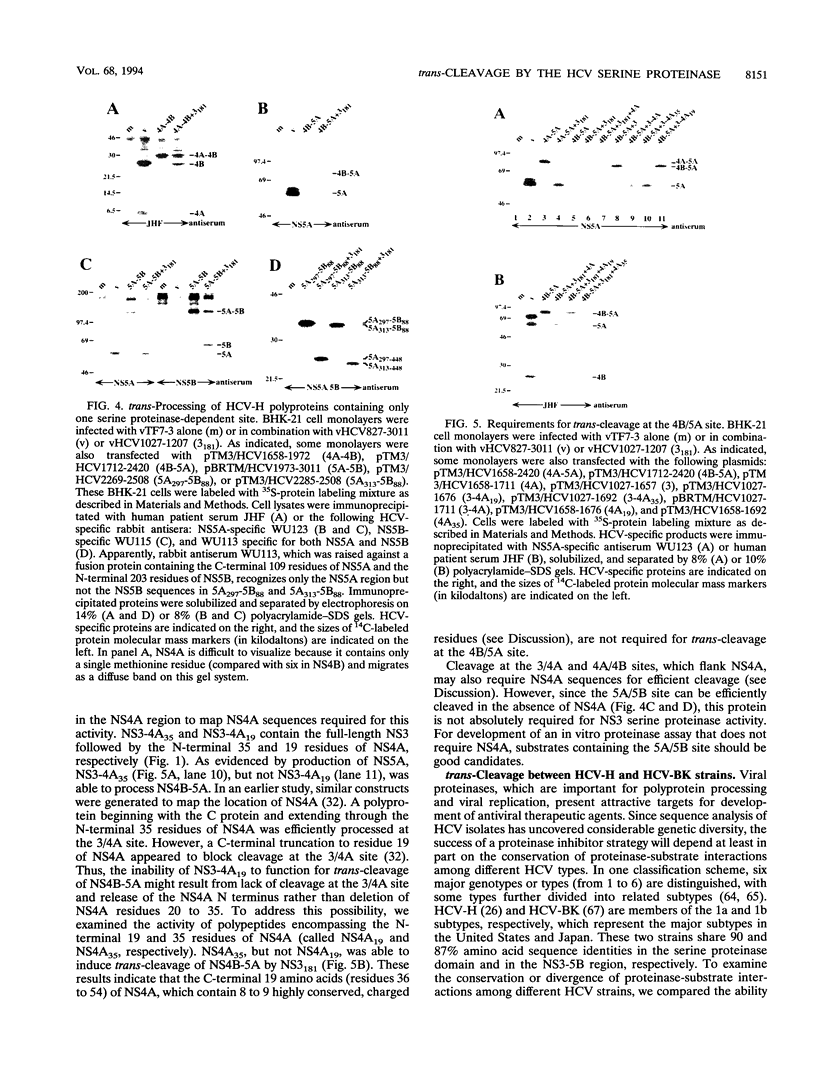
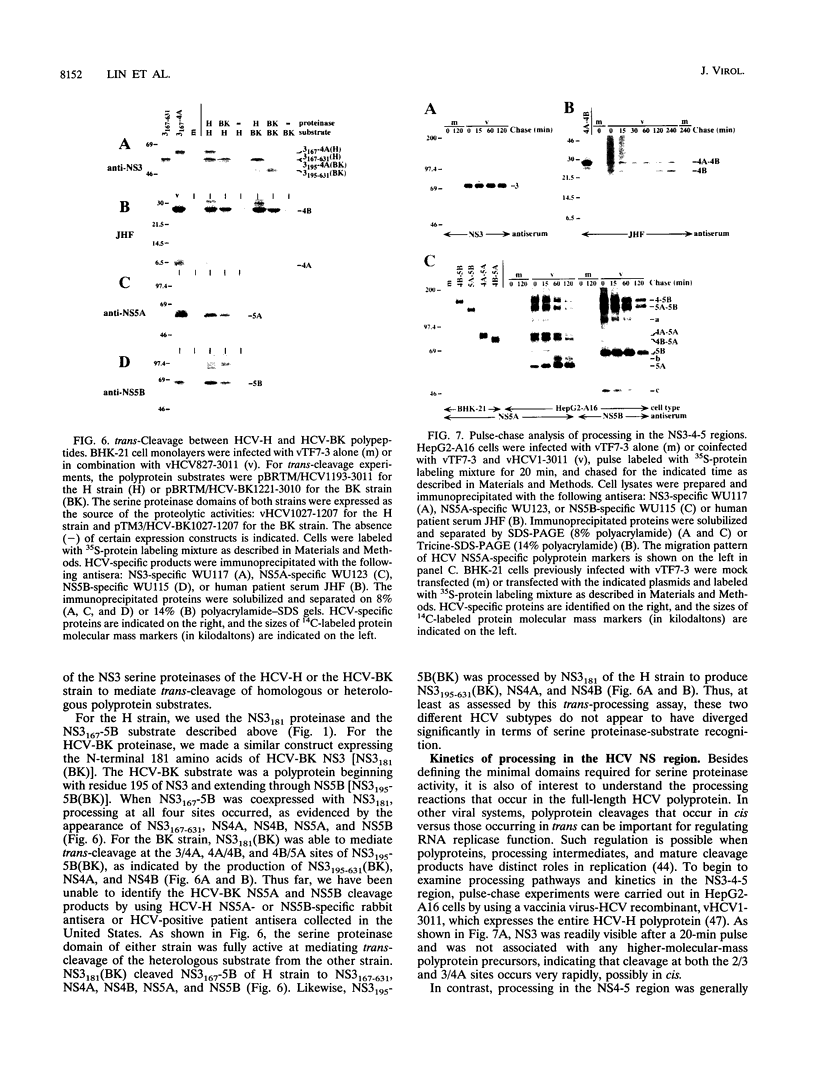
The hepatitis C virus H strain (HCV-H) polyprotein is cleaved to produce at least 10 distinct products, in the order of NH2-C-E1-E2-p7-NS2-NS3-NS4A-NS4B-NS5A-NS5B -COOH. An HCV-encoded serine proteinase activity in NS3 is required for cleavage at four sites in the nonstructural region (3/4A, 4A/4B, 4B/5A, and 5A/5B). In this report, the HCV-H serine proteinase domain (the N-terminal 181 residues of NS3) was tested for its ability to mediate trans-processing at these four sites. By using an NS3-5B substrate with an inactivated serine proteinase domain, trans-cleavage was observed at all sites except for the 3/4A site. Deletion of the inactive proteinase domain led to efficient trans-processing at the 3/4A site. Smaller NS4A-4B and NS5A-5B substrates were processed efficiently in trans; however, cleavage of an NS4B-5A substrate occurred only when the serine proteinase domain was coexpressed with NS4A. Only the N-terminal 35 amino acids of NS4A were required for this activity. Thus, while NS4A appears to be absolutely required for trans-cleavage at the 4B/5A site, it is not an essential cofactor for serine proteinase activity. To begin to examine the conservation (or divergence) of serine proteinase-substrate interactions during HCV evolution, we demonstrated that similar trans-processing occurred when the proteinase domains and substrates were derived from two different HCV subtypes. These results are encouraging for the development of broadly effective HCV serine proteinase inhibitors as antiviral agents. Finally, the kinetics of processing in the nonstructural region was examined by pulse-chase analysis. NS3-containing precursors were absent, indicating that the 2/3 and 3/4A cleavages occur rapidly. In contrast, processing of the NS4A-5B region appeared to involve multiple pathways, and significant quantities of various polyprotein intermediates were observed. NS5B, the putative RNA polymerase, was found to be significantly less stable than the other mature cleavage products. This instability appeared to be an inherent property of NS5B and did not depend on expression of other viral polypeptides, including the HCV-encoded proteinases.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alter M. J., Hadler S. C., Judson F. N., Mares A., Alexander W. J., Hu P. Y., Miller J. K., Moyer L. A., Fields H. A., Bradley D. W. Risk factors for acute non-A, non-B hepatitis in the United States and association with hepatitis C virus infection. JAMA. 1990 Nov 7;264(17):2231–2235. [PubMed] [Google Scholar]
- Amberg S. M., Nestorowicz A., McCourt D. W., Rice C. M. NS2B-3 proteinase-mediated processing in the yellow fever virus structural region: in vitro and in vivo studies. J Virol. 1994 Jun;68(6):3794–3802. doi: 10.1128/jvi.68.6.3794-3802.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias C. F., Preugschat F., Strauss J. H. Dengue 2 virus NS2B and NS3 form a stable complex that can cleave NS3 within the helicase domain. Virology. 1993 Apr;193(2):888–899. doi: 10.1006/viro.1993.1198. [DOI] [PubMed] [Google Scholar]
- Bachmair A., Finley D., Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986 Oct 10;234(4773):179–186. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- Bartenschlager R., Ahlborn-Laake L., Mous J., Jacobsen H. Kinetic and structural analyses of hepatitis C virus polyprotein processing. J Virol. 1994 Aug;68(8):5045–5055. doi: 10.1128/jvi.68.8.5045-5055.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartenschlager R., Ahlborn-Laake L., Mous J., Jacobsen H. Nonstructural protein 3 of the hepatitis C virus encodes a serine-type proteinase required for cleavage at the NS3/4 and NS4/5 junctions. J Virol. 1993 Jul;67(7):3835–3844. doi: 10.1128/jvi.67.7.3835-3844.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredenbeek P. J., Frolov I., Rice C. M., Schlesinger S. Sindbis virus expression vectors: packaging of RNA replicons by using defective helper RNAs. J Virol. 1993 Nov;67(11):6439–6446. doi: 10.1128/jvi.67.11.6439-6446.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockman W. W., Nathans D. The isolation of simian virus 40 variants with specifically altered genomes. Proc Natl Acad Sci U S A. 1974 Mar;71(3):942–946. doi: 10.1073/pnas.71.3.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukh J., Purcell R. H., Miller R. H. At least 12 genotypes of hepatitis C virus predicted by sequence analysis of the putative E1 gene of isolates collected worldwide. Proc Natl Acad Sci U S A. 1993 Sep 1;90(17):8234–8238. doi: 10.1073/pnas.90.17.8234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers T. J., Grakoui A., Rice C. M. Processing of the yellow fever virus nonstructural polyprotein: a catalytically active NS3 proteinase domain and NS2B are required for cleavages at dibasic sites. J Virol. 1991 Nov;65(11):6042–6050. doi: 10.1128/jvi.65.11.6042-6050.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers T. J., Hahn C. S., Galler R., Rice C. M. Flavivirus genome organization, expression, and replication. Annu Rev Microbiol. 1990;44:649–688. doi: 10.1146/annurev.mi.44.100190.003245. [DOI] [PubMed] [Google Scholar]
- Chambers T. J., Nestorowicz A., Amberg S. M., Rice C. M. Mutagenesis of the yellow fever virus NS2B protein: effects on proteolytic processing, NS2B-NS3 complex formation, and viral replication. J Virol. 1993 Nov;67(11):6797–6807. doi: 10.1128/jvi.67.11.6797-6807.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers T. J., Weir R. C., Grakoui A., McCourt D. W., Bazan J. F., Fletterick R. J., Rice C. M. Evidence that the N-terminal domain of nonstructural protein NS3 from yellow fever virus is a serine protease responsible for site-specific cleavages in the viral polyprotein. Proc Natl Acad Sci U S A. 1990 Nov;87(22):8898–8902. doi: 10.1073/pnas.87.22.8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P. J., Lin M. H., Tai K. F., Liu P. C., Lin C. J., Chen D. S. The Taiwanese hepatitis C virus genome: sequence determination and mapping the 5' termini of viral genomic and antigenomic RNA. Virology. 1992 May;188(1):102–113. doi: 10.1016/0042-6822(92)90739-c. [DOI] [PubMed] [Google Scholar]
- Choo Q. L., Kuo G., Weiner A. J., Overby L. R., Bradley D. W., Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989 Apr 21;244(4902):359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- Choo Q. L., Richman K. H., Han J. H., Berger K., Lee C., Dong C., Gallegos C., Coit D., Medina-Selby R., Barr P. J. Genetic organization and diversity of the hepatitis C virus. Proc Natl Acad Sci U S A. 1991 Mar 15;88(6):2451–2455. doi: 10.1073/pnas.88.6.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A., DiGiuseppe J. A., Bercovich B., Orian A., Richter J. D., Schwartz A. L., Brodeur G. M. Degradation of nuclear oncoproteins by the ubiquitin system in vitro. Proc Natl Acad Sci U S A. 1991 Jan 1;88(1):139–143. doi: 10.1073/pnas.88.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collett M. S. Molecular genetics of pestiviruses. Comp Immunol Microbiol Infect Dis. 1992 Jul;15(3):145–154. doi: 10.1016/0147-9571(92)90087-8. [DOI] [PubMed] [Google Scholar]
- Collett M. S., Wiskerchen M., Welniak E., Belzer S. K. Bovine viral diarrhea virus genomic organization. Arch Virol Suppl. 1991;3:19–27. doi: 10.1007/978-3-7091-9153-8_3. [DOI] [PubMed] [Google Scholar]
- Eckart M. R., Selby M., Masiarz F., Lee C., Berger K., Crawford K., Kuo C., Kuo G., Houghton M., Choo Q. L. The hepatitis C virus encodes a serine protease involved in processing of the putative nonstructural proteins from the viral polyprotein precursor. Biochem Biophys Res Commun. 1993 Apr 30;192(2):399–406. doi: 10.1006/bbrc.1993.1429. [DOI] [PubMed] [Google Scholar]
- Failla C., Tomei L., De Francesco R. Both NS3 and NS4A are required for proteolytic processing of hepatitis C virus nonstructural proteins. J Virol. 1994 Jun;68(6):3753–3760. doi: 10.1128/jvi.68.6.3753-3760.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falgout B., Pethel M., Zhang Y. M., Lai C. J. Both nonstructural proteins NS2B and NS3 are required for the proteolytic processing of dengue virus nonstructural proteins. J Virol. 1991 May;65(5):2467–2475. doi: 10.1128/jvi.65.5.2467-2475.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkner F. G., Moss B. Escherichia coli gpt gene provides dominant selection for vaccinia virus open reading frame expression vectors. J Virol. 1988 Jun;62(6):1849–1854. doi: 10.1128/jvi.62.6.1849-1854.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinstone S. M., Alter H. J., Dienes H. P., Shimizu Y., Popper H., Blackmore D., Sly D., London W. T., Purcell R. H. Non-A, non-B hepatitis in chimpanzees and marmosets. J Infect Dis. 1981 Dec;144(6):588–598. doi: 10.1093/infdis/144.6.588. [DOI] [PubMed] [Google Scholar]
- Fuerst T. R., Niles E. G., Studier F. W., Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci U S A. 1986 Nov;83(21):8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A. E., Donchenko A. P., Koonin E. V., Blinov V. M. N-terminal domains of putative helicases of flavi- and pestiviruses may be serine proteases. Nucleic Acids Res. 1989 May 25;17(10):3889–3897. doi: 10.1093/nar/17.10.3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grakoui A., McCourt D. W., Wychowski C., Feinstone S. M., Rice C. M. A second hepatitis C virus-encoded proteinase. Proc Natl Acad Sci U S A. 1993 Nov 15;90(22):10583–10587. doi: 10.1073/pnas.90.22.10583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grakoui A., McCourt D. W., Wychowski C., Feinstone S. M., Rice C. M. Characterization of the hepatitis C virus-encoded serine proteinase: determination of proteinase-dependent polyprotein cleavage sites. J Virol. 1993 May;67(5):2832–2843. doi: 10.1128/jvi.67.5.2832-2843.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grakoui A., Wychowski C., Lin C., Feinstone S. M., Rice C. M. Expression and identification of hepatitis C virus polyprotein cleavage products. J Virol. 1993 Mar;67(3):1385–1395. doi: 10.1128/jvi.67.3.1385-1395.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hijikata M., Kato N., Ootsuyama Y., Nakagawa M., Shimotohno K. Gene mapping of the putative structural region of the hepatitis C virus genome by in vitro processing analysis. Proc Natl Acad Sci U S A. 1991 Jul 1;88(13):5547–5551. doi: 10.1073/pnas.88.13.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hijikata M., Mizushima H., Akagi T., Mori S., Kakiuchi N., Kato N., Tanaka T., Kimura K., Shimotohno K. Two distinct proteinase activities required for the processing of a putative nonstructural precursor protein of hepatitis C virus. J Virol. 1993 Aug;67(8):4665–4675. doi: 10.1128/jvi.67.8.4665-4675.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hijikata M., Mizushima H., Tanji Y., Komoda Y., Hirowatari Y., Akagi T., Kato N., Kimura K., Shimotohno K. Proteolytic processing and membrane association of putative nonstructural proteins of hepatitis C virus. Proc Natl Acad Sci U S A. 1993 Nov 15;90(22):10773–10777. doi: 10.1073/pnas.90.22.10773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houghton M., Weiner A., Han J., Kuo G., Choo Q. L. Molecular biology of the hepatitis C viruses: implications for diagnosis, development and control of viral disease. Hepatology. 1991 Aug;14(2):381–388. [PubMed] [Google Scholar]
- Hruby D. E., Guarino L. A., Kates J. R. Vaccinia virus replication. I. Requirement for the host-cell nucleus. J Virol. 1979 Feb;29(2):705–715. doi: 10.1128/jvi.29.2.705-715.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inchauspe G., Zebedee S., Lee D. H., Sugitani M., Nasoff M., Prince A. M. Genomic structure of the human prototype strain H of hepatitis C virus: comparison with American and Japanese isolates. Proc Natl Acad Sci U S A. 1991 Nov 15;88(22):10292–10296. doi: 10.1073/pnas.88.22.10292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato N., Hijikata M., Ootsuyama Y., Nakagawa M., Ohkoshi S., Sugimura T., Shimotohno K. Molecular cloning of the human hepatitis C virus genome from Japanese patients with non-A, non-B hepatitis. Proc Natl Acad Sci U S A. 1990 Dec;87(24):9524–9528. doi: 10.1073/pnas.87.24.9524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolykhalov A. A., Agapov E. V., Rice C. M. Specificity of the hepatitis C virus NS3 serine protease: effects of substitutions at the 3/4A, 4A/4B, 4B/5A, and 5A/5B cleavage sites on polyprotein processing. J Virol. 1994 Nov;68(11):7525–7533. doi: 10.1128/jvi.68.11.7525-7533.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo G., Choo Q. L., Alter H. J., Gitnick G. L., Redeker A. G., Purcell R. H., Miyamura T., Dienstag J. L., Alter M. J., Stevens C. E. An assay for circulating antibodies to a major etiologic virus of human non-A, non-B hepatitis. Science. 1989 Apr 21;244(4902):362–364. doi: 10.1126/science.2496467. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Laskey R. A., Mills A. D. Quantitative film detection of 3H and 14C in polyacrylamide gels by fluorography. Eur J Biochem. 1975 Aug 15;56(2):335–341. doi: 10.1111/j.1432-1033.1975.tb02238.x. [DOI] [PubMed] [Google Scholar]
- Lemm J. A., Rümenapf T., Strauss E. G., Strauss J. H., Rice C. M. Polypeptide requirements for assembly of functional Sindbis virus replication complexes: a model for the temporal regulation of minus- and plus-strand RNA synthesis. EMBO J. 1994 Jun 15;13(12):2925–2934. doi: 10.1002/j.1460-2075.1994.tb06587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C., Amberg S. M., Chambers T. J., Rice C. M. Cleavage at a novel site in the NS4A region by the yellow fever virus NS2B-3 proteinase is a prerequisite for processing at the downstream 4A/4B signalase site. J Virol. 1993 Apr;67(4):2327–2335. doi: 10.1128/jvi.67.4.2327-2335.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C., Chambers T. J., Rice C. M. Mutagenesis of conserved residues at the yellow fever virus 3/4A and 4B/5 dibasic cleavage sites: effects on cleavage efficiency and polyprotein processing. Virology. 1993 Feb;192(2):596–604. doi: 10.1006/viro.1993.1076. [DOI] [PubMed] [Google Scholar]
- Lin C., Lindenbach B. D., Prágai B. M., McCourt D. W., Rice C. M. Processing in the hepatitis C virus E2-NS2 region: identification of p7 and two distinct E2-specific products with different C termini. J Virol. 1994 Aug;68(8):5063–5073. doi: 10.1128/jvi.68.8.5063-5073.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobigs M. Proteolytic processing of a Murray Valley encephalitis virus non-structural polyprotein segment containing the viral proteinase: accumulation of a NS3-4A precursor which requires mature NS3 for efficient processing. J Gen Virol. 1992 Sep;73(Pt 9):2305–2312. doi: 10.1099/0022-1317-73-9-2305. [DOI] [PubMed] [Google Scholar]
- Mackett M., Smith G. L. Vaccinia virus expression vectors. J Gen Virol. 1986 Oct;67(Pt 10):2067–2082. doi: 10.1099/0022-1317-67-10-2067. [DOI] [PubMed] [Google Scholar]
- Nestorowicz A., Chambers T. J., Rice C. M. Mutagenesis of the yellow fever virus NS2A/2B cleavage site: effects on proteolytic processing, viral replication, and evidence for alternative processing of the NS2A protein. Virology. 1994 Feb 15;199(1):114–123. doi: 10.1006/viro.1994.1103. [DOI] [PubMed] [Google Scholar]
- Okamoto H., Kojima M., Sakamoto M., Iizuka H., Hadiwandowo S., Suwignyo S., Miyakawa Y., Mayumi M. The entire nucleotide sequence and classification of a hepatitis C virus isolate of a novel genotype from an Indonesian patient with chronic liver disease. J Gen Virol. 1994 Mar;75(Pt 3):629–635. doi: 10.1099/0022-1317-75-3-629. [DOI] [PubMed] [Google Scholar]
- Okamoto H., Kurai K., Okada S., Yamamoto K., Lizuka H., Tanaka T., Fukuda S., Tsuda F., Mishiro S. Full-length sequence of a hepatitis C virus genome having poor homology to reported isolates: comparative study of four distinct genotypes. Virology. 1992 May;188(1):331–341. doi: 10.1016/0042-6822(92)90762-e. [DOI] [PubMed] [Google Scholar]
- Okamoto H., Okada S., Sugiyama Y., Kurai K., Iizuka H., Machida A., Miyakawa Y., Mayumi M. Nucleotide sequence of the genomic RNA of hepatitis C virus isolated from a human carrier: comparison with reported isolates for conserved and divergent regions. J Gen Virol. 1991 Nov;72(Pt 11):2697–2704. doi: 10.1099/0022-1317-72-11-2697. [DOI] [PubMed] [Google Scholar]
- Preugschat F., Strauss J. H. Processing of nonstructural proteins NS4A and NS4B of dengue 2 virus in vitro and in vivo. Virology. 1991 Dec;185(2):689–697. doi: 10.1016/0042-6822(91)90540-r. [DOI] [PubMed] [Google Scholar]
- Preugschat F., Yao C. W., Strauss J. H. In vitro processing of dengue virus type 2 nonstructural proteins NS2A, NS2B, and NS3. J Virol. 1990 Sep;64(9):4364–4374. doi: 10.1128/jvi.64.9.4364-4374.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice C. M., Levis R., Strauss J. H., Huang H. V. Production of infectious RNA transcripts from Sindbis virus cDNA clones: mapping of lethal mutations, rescue of a temperature-sensitive marker, and in vitro mutagenesis to generate defined mutants. J Virol. 1987 Dec;61(12):3809–3819. doi: 10.1128/jvi.61.12.3809-3819.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice C. M., Strauss J. H. Association of sindbis virion glycoproteins and their precursors. J Mol Biol. 1982 Jan 15;154(2):325–348. doi: 10.1016/0022-2836(82)90067-5. [DOI] [PubMed] [Google Scholar]
- Sakamoto M., Akahane Y., Tsuda F., Tanaka T., Woodfield D. G., Okamoto H. Entire nucleotide sequence and characterization of a hepatitis C virus of genotype V/3a. J Gen Virol. 1994 Jul;75(Pt 7):1761–1768. doi: 10.1099/0022-1317-75-7-1761. [DOI] [PubMed] [Google Scholar]
- Schägger H., von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987 Nov 1;166(2):368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- Simmonds P., Holmes E. C., Cha T. A., Chan S. W., McOmish F., Irvine B., Beall E., Yap P. L., Kolberg J., Urdea M. S. Classification of hepatitis C virus into six major genotypes and a series of subtypes by phylogenetic analysis of the NS-5 region. J Gen Virol. 1993 Nov;74(Pt 11):2391–2399. doi: 10.1099/0022-1317-74-11-2391. [DOI] [PubMed] [Google Scholar]
- Simmonds P., Smith D. B., McOmish F., Yap P. L., Kolberg J., Urdea M. S., Holmes E. C. Identification of genotypes of hepatitis C virus by sequence comparisons in the core, E1 and NS-5 regions. J Gen Virol. 1994 May;75(Pt 5):1053–1061. doi: 10.1099/0022-1317-75-5-1053. [DOI] [PubMed] [Google Scholar]
- Strauss J. H., Strauss E. G. The alphaviruses: gene expression, replication, and evolution. Microbiol Rev. 1994 Sep;58(3):491–562. doi: 10.1128/mr.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamizawa A., Mori C., Fuke I., Manabe S., Murakami S., Fujita J., Onishi E., Andoh T., Yoshida I., Okayama H. Structure and organization of the hepatitis C virus genome isolated from human carriers. J Virol. 1991 Mar;65(3):1105–1113. doi: 10.1128/jvi.65.3.1105-1113.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T., Kato N., Nakagawa M., Ootsuyama Y., Cho M. J., Nakazawa T., Hijikata M., Ishimura Y., Shimotohno K. Molecular cloning of hepatitis C virus genome from a single Japanese carrier: sequence variation within the same individual and among infected individuals. Virus Res. 1992 Apr;23(1-2):39–53. doi: 10.1016/0168-1702(92)90066-i. [DOI] [PubMed] [Google Scholar]
- Tomei L., Failla C., Santolini E., De Francesco R., La Monica N. NS3 is a serine protease required for processing of hepatitis C virus polyprotein. J Virol. 1993 Jul;67(7):4017–4026. doi: 10.1128/jvi.67.7.4017-4026.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wengler G., Czaya G., Färber P. M., Hegemann J. H. In vitro synthesis of West Nile virus proteins indicates that the amino-terminal segment of the NS3 protein contains the active centre of the protease which cleaves the viral polyprotein after multiple basic amino acids. J Gen Virol. 1991 Apr;72(Pt 4):851–858. doi: 10.1099/0022-1317-72-4-851. [DOI] [PubMed] [Google Scholar]
- Wiskerchen M., Collett M. S. Pestivirus gene expression: protein p80 of bovine viral diarrhea virus is a proteinase involved in polyprotein processing. Virology. 1991 Sep;184(1):341–350. doi: 10.1016/0042-6822(91)90850-b. [DOI] [PubMed] [Google Scholar]
- Zhang L., Mohan P. M., Padmanabhan R. Processing and localization of Dengue virus type 2 polyprotein precursor NS3-NS4A-NS4B-NS5. J Virol. 1992 Dec;66(12):7549–7554. doi: 10.1128/jvi.66.12.7549-7554.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]