Abstract
The mechanism of bacterial gliding motility (active movement over surfaces without the aid of flagella) is not known. A large number of nonmotile mutants of the gliding bacterium Flavobacterium johnsoniae (Cytophaga johnsonae) have been previously isolated, and genetic techniques to analyze these mutants have recently been developed. We complemented a nonmotile mutant of F. johnsoniae (UW102-09) with a library of wild-type DNA by using the shuttle cosmid pCP17. The complementing plasmid (pCP100) contained an insert of 13 kbp, and restored motility to 4 of 61 independently isolated nonmotile mutants. A 1.3-kbp fragment that encompassed a single ORF, gldA, complemented all four mutants. Disruption of the chromosomal copy of gldA in wild-type F. johnsoniae UW101 eliminated gliding motility. The predicted protein produced by gldA has strong sequence similarity to ATP binding cassette transport proteins.
Bacteria from many of the branches of the eubacterial phylogenetic tree are able to move over surfaces by a process known as gliding motility (1). Gliding bacteria lack flagella or other obvious motility organelles. Numerous models have been proposed to explain gliding motility, but the mechanism responsible for cell movement is not known (2, 3). Flavobacterium johnsoniae [formerly Cytophaga johnsonae (4)] is a Gram-negative bacterium that exhibits rapid gliding motility (3). The ultrastructure and biochemistry of cells of F. johnsoniae have been the subject of numerous studies aimed at determining the mechanism of bacterial gliding motility (5–7). In addition, a large number of nonmotile mutants of F. johnsoniae have been isolated (8, 9). Unlike wild-type F. johnsoniae, these mutants form nonspreading colonies under all conditions of cultivation. They also lack the single cell movements that are observed with wild-type cells. Techniques to genetically manipulate F. johnsoniae and related bacteria were recently developed (10, 11). Here we report the use of these techniques to clone and analyze a gene that is required for F. johnsoniae gliding motility.
MATERIALS AND METHODS
Bacterial and Bacteriophage Strains, Plasmids, and Growth Conditions.
F. johnsoniae UW101 (ATCC 17061) was the wild-type strain used in these studies and all mutants were derived from this strain. The 61 nonmotile mutants of F. johnsoniae (obtained from J. Pate) were previously described (8, 9). The strain designations for each of these nonmotile mutants carry the prefix ‘UW102-’. The strain designations are UW102-9, -15, -21, -25, -33, -34, -39, -40, -41, -42, -45, -48, -50, -51, -52, -53, -55, -56, -57, -58, -61, -64, -66, -68, -69, -73, -75, -77, -78, -80, -81, -85, -86, -90, -91, -92, -94, -95, -96, -97, -98, -99, -100, -101, -103, -106, -107, -108, -137, -140, -141, -146, -149, -154, -155, -156, -300, -301, -302, -345, and -348. The bacteriophage active against F. johnsoniae that were used in this study (φCj1, φCj7, φCj13, φCj23, φCj28, φCj29, φCj42, φCj48, and φCj54) have been described (8, 9, 12). The Escherichia coli strains used were DH5αMCR (GIBCO/BRL), HB101(13), and S17-1 (14). E. coli strains were grown in Luria–Bertani medium at 37°C, and F. johnsoniae strains were grown in casitone yeast extract medium at 30°C, as described (10). When we wished to observe colony-spreading, F. johnsoniae was grown on PY2 medium (2 g peptone/0.5 g yeast extract/10 g agar/liter, pH 7.3) at 25°C. Antibiotics were used at the following concentrations when needed: ampicillin (100 μg/ml), chloramphenicol (30 μg/ml), erythromycin (Em) (100 μg/ml), tetracycline (15 μg/ml), kanamycin (30 μg/ml), and streptomycin (30 μg/ml). Plasmids used in this study are listed in Table 1 (10, 15–18).
Table 1.
Plasmids used in this study
| Plasmid | Description | Ref. |
|---|---|---|
| pUC119 | pUC, Apr | 15 |
| pBC SK+ | ColEl, Cmr | Stratagene |
| pBC KS+ | ColEl, Cmr | Stratagene |
| pCR-Script | ColEl, Apr | Stratagene |
| pUC4K | pUC, AprKmr | Pharmacia |
| R702 | Helper plasmid for triparental conjugation, KmrSmrTcr | 16 |
| pCP11 | E. coli–F. johnsoniae shuttle plasmid, pUC ori, AprEmr* | 10 |
| pCP17 | E. coli–F. johnsoniae shuttle cosmid, pUC ori, AprTcrEmr* | 10 |
| pLYL03 | Bacteroides–Flavobacterium suicide vector used to make chromosomal insertions, pUC ori, AprEmr* | 17 |
| pLYL001 | Bacteroides–Flavobacterium suicide vector, pUC ori, AprTcr* | 18 |
| pCP23 | E. coli–F. johnsoniae shuttle plasmid, AprTcr* | This study |
| pCP100 | Cosmid clone complementing F. johnsoniae UW 102-9, AprEmr* | This study |
| pSAR2 | 4.5-kbp XbaI/ScaI fragment of pCP100 containing gldA cloned into the SmaI and XbaI sites of pUC119, Apr | This study |
| pCP25 | 4.5-kbp XbaI/SacI fragment of pSAR2 containing gldA cloned into the XbaI and SacI sites of pCP11, AprEmr* | This study |
| pCP105 | 2-kbp XbaI/BamHI fragment of pCP100 cloned in pCP11, AprEmr* | This study |
| pCP104 | 5-kbp BamHI/SalI fragment of pCP100 cloned in pCP11, AprEmr* | This study |
| pMM114 | HindIII/BamHI fragment of gldA cloned in pLYL03, AprEmr* | This study |
| pSA10 | 1.3-kbp PCR product containing gldA cloned into the SrfI site of pCR-Script, Apr | This study |
| pSA11 | 1.3-kbp SacI/XbaI fragment of pSA10 containing gldA cloned in pCP11, AprEmr* | This study |
| pSA20 | 1.3-kbp SacI/XbaI fragment of pSA10 containing gldA cloned into pBC SK+, Cmr | This study |
| pSA21 | 1.3-kbp XbaI/KpnI fragment of pSA20 containing gldA cloned into pCP23, AprTcr* | This study |
| pDH107 | 605-bp ClaI/HindIII fragment of orf1 cloned into the ClaI and HindIII sites of pUC4K, Apr | This study |
| pDH109-5 | 1.5-kbp PstI fragment of pDH107 which contains the entire 605 bp ClaI/HindIII fragment of orf1 and 740 bp of flanking DNA from the kanamycin resistance gene cloned into pLYL03, Apr Emr* | This study |
Unless indicated otherwise, antibiotic resistances are those expressed in E. coli.
Antibiotic resistance is expressed in F. johnsoniae but not in E. coli.
Construction of a Library of Wild-Type F. johnsoniae DNA.
F. johnsoniae DNA was partially digested with Sau3AI and ligated with the shuttle cosmid pCP17 that had been digested with BamHI and treated with alkaline phosphatase. The DNA was packaged into λ phage particles and introduced into E. coli DH5αMCR. Approximately 10,000 colonies were pooled and mixed with an equal amount of E. coli HB101 carrying the helper plasmid R702, and the cosmids were transferred by triparental conjugation into F. johnsoniae UW102-9. The transconjugants were plated on PY2 medium containing 100 μg/ml Em and incubated for 2–3 days at 25°C.
Subcloning of pCP100 and Complementation of Nonmotile Mutants.
The plasmid pCP100, which complemented the nonmotile mutant UW102-9, was subcloned by using standard procedures (19). Plasmid DNA was transferred to nonmotile mutants by conjugation or electroporation as described (10), and the transformants were plated on PY2 medium containing 100 μg/ml Em and observed for colony spreading. The minimal complementing fragment containing gldA was obtained by PCR amplification using the primers 5′-ATTCGAGCTCTTCAGAAGTATAACCGATG-3′ (SacI site added) and 5′-ATTATCTAGATGCTTGGCAAATATAACAC-3′ (XbaI site added) and using pSAR2 as the template. pSAR2 contains the 4.5-kbp XbaI/ScaI fragment of pCP100 inserted between the XbaI and SmaI sites of pUC119. The 1.3-kbp PCR product was cloned into the SrfI site of PCR-Script (Stratagene) to generate pSA10. pSA10 was digested with SacI and XbaI and the 1.3-kbp fragment was ligated into pCP11, which had been digested with the same enzymes, to generate pSA11.
Nucleic Acid Sequencing.
Nucleic acid sequencing was performed by the dideoxy nucleotide procedure by using a combination of manual and automated (Applied Biosystems) sequencing systems. Sequences were analyzed with the macvector and assemblylign software (Oxford Molecular Group, Campbell, CA), and comparisons to database sequences were made using the blast (20) and fasta (21) algorithms. Pairwise sequence alignments were performed with the lalign program (22).
Disruption of gldA.
To disrupt the chromosomal copy of gldA in wild-type F. johnsoniae, we cloned the 438-bp HindIII/BamHI fragment that lies entirely within gldA into the F. johnsoniae suicide vector pLYL03, to generate pMM114. This fragment of gldA extends from nucleotide 47 to nucleotide 485 of the predicted 897-bp coding sequence. pMM114 was introduced into F. johnsoniae UW101 by conjugation from E. coli S17-1. Em-resistant colonies arose after insertion of pMM114 into the chromosomal copy of gldA, to generate CJ101-288 (verified by Southern blot hybridization).
Construction of pCP23.
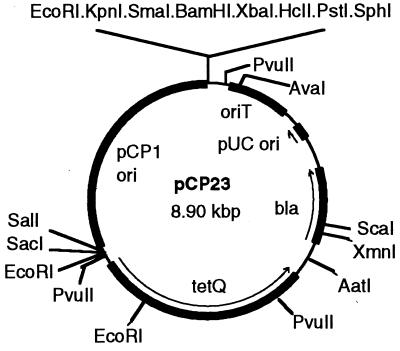
pSA11, which confers Em resistance on F. johnsoniae, could not be used to complement the gldA knockout mutant CJ101-288 because this mutant is already Em resistant. For this reason, we identified an additional antibiotic resistance gene that is expressed in F. johnsoniae (tetQ) and constructed a F. johnsoniae–E. coli shuttle plasmid (pCP23) that confers tetracycline resistance on F. johnsoniae. pCP11 was digested with PstI and ScaI and the ends were filled in with DNA polymerase Klenow fragment. The 6.4-kbp fragment containing the pCP1 origin of replication (10) and the ermF gene was ligated to itself to generate pCP18. pCP18 replicates in F. johnsoniae but not in E. coli. The 0.75-kbp PvuII/SmaI fragment was removed from pCP18 to generate pCP19. pCP19 was digested with KpnI and SacI, to remove ermF, and was ligated with pLYL001, which had been digested with the same enzymes, to generate pCP23 (Fig. 1). pCP23 confers resistance to 10–20 μg/ml tetracycline on F. johnsoniae and confers resistance to 100 μg/ml ampicillin on E. coli.
Figure 1.
Map of the F. johnsoniae–E. coli shuttle plasmid pCP23.
Complementation of the gldA Mutants CJ101-288 and CJ557.
pSA10 was digested with XbaI and SacI and the 1.3-kbp fragment was inserted into pBC KS+ that had been cut with the same enzymes, to generate pSA20. The 1.3-kbp XbaI/KpnI fragment from pSA20 containing gldA was introduced into pCP23 to generate pSA21. pSA21 was introduced into CJ101-288 and CJ557 (Tn4351-induced gldA mutant) by conjugation to test for complementation of the gldA mutants.
Disruption of orf1.
To disrupt the chromosomal copy of orf1 in wild-type cells, we cloned a 605-bp ClaI/HindIII fragment that lies entirely within orf1 into pUC4K, to generate pDH107. This fragment extends from nucleotide 155 to nucleotide 760 of the predicted orf1 coding region. It was then transferred into pLYL03 as a PstI fragment to generate pDH109-5. pDH109-5 was introduced into F. johnsoniae UW101 by conjugation from E. coli S17-1. Em-resistant colonies arose following insertion of pDH109-5 into the chromosomal copy of orf1 (verified by Southern blot hybridization) to generate CJ101-289.
Microscopic Observations of Cell Movement and Bead Movement.
Wild-type and mutant cells of F. johnsoniae were examined for movement over glass and agar surfaces by phase contrast microscopy on a heated stage at 25°C by using two different assays. Cells to be observed were grown on PY2 agar medium overnight at 25°C, scraped off of the agar, and suspended in 10 mM Tris (pH 7.3)/8 mM CaCl2 (TC buffer) at a density of about 108 cells per ml. To observe movement over glass, a drop of the cell suspension was placed on a microscope slide, covered with an oxygen-permeable membrane (Yellow Springs Instruments) that functioned as a coverslip, and examined by phase contrast microscopy. To observe movement of cells on agar, 5 μl of the cell suspension in TC buffer was spotted onto a layer of PY2 agar on a microscope slide. The spot was allowed to dry partially (less than 5 min), after which it was covered with an oxygen-permeable membrane. After 1-h incubation at 25°C in a moist chamber, cells were examined by time lapse videomicroscopy for movement. The videotape was played back at 60× the recording speed to allow detection of slow or intermittent movements.
We also examined the ability of cells to bind to and propel 0.5 μm polystyrene latex spheres (Seradyn, Indianapolis, IN). Cells were grown to mid-exponential phase in casitone yeast extract broth. Latex spheres in dH2O were added to a final concentration of approximately 0.03 g/liter. The cell suspension (0.05 ml) was spotted on a microscope slide, covered with an O2-permeable membrane, and examined by phase contrast microscopy.
Measurements of Phage Sensitivity.
Sensitivity to F. johnsoniae phage was determined essentially as described by spotting 1–10 μl of phage lysates (2 × 107 phage/ml) onto lawns of cells in overlay agar (9). The plates were incubated for 24 h at 25°C to observe lysis.
RESULTS AND DISCUSSION
Complementation of Nonmotile Mutants.
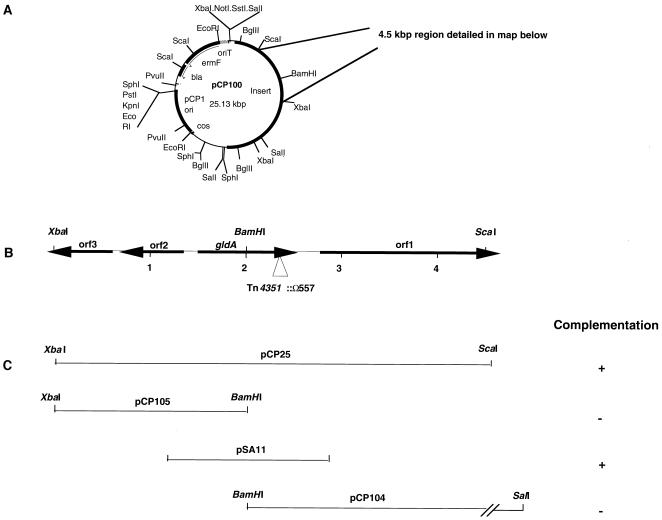
To identify and clone genes involved in gliding motility, we constructed a library of wild-type F. johnsoniae DNA in the F. johnsoniae–E. coli shuttle cosmid pCP17. The cosmid clones were transferred to the nonmotile mutant UW102-9 and we identified one colony that exhibited spreading. The cells of this colony carried the cosmid clone pCP100 (Fig. 2A)that had a 13-kbp insert of F. johnsoniae DNA. We introduced pCP100 into UW102-9 by electroporation and verified that pCP100 complemented the motility defect of this mutant. All of the Em-resistant colonies carrying pCP100 exhibited spreading, and individual cells exhibited rapid movement over glass or agar surfaces when examined by phase contrast microscopy. We introduced pCP100 into each of 61 independently isolated nonmotile mutants (listed in Materials and Methods). pCP100 complemented UW102-15, UW102-107, and UW102-146, in addition to UW102-9, but did not complement the other 57 mutants.
Figure 2.
Map of the region of DNA surrounding gldA. (A) Map of pCP100. pCP100 contains a 13-kbp fragment of F. johnsoniae DNA inserted into the F. johnsoniae–E. coli shuttle cosmid pCP17. (B) Map of the 4.5-kbp XbaI/ScaI fragment that contains gldA. The site of the Tn4351 insert in CJ557 is indicated by a triangle. (C) Complementation of motility mutants UW102-9, UW102-15, UW102-107, and UW102-146 by fragments cloned into pCP11.
Subcloning and Sequencing of gldA.
We cloned a 4.5-kbp XbaI/ScaI fragment of pCP100 into pCP11 to generate pCP25 (Fig. 2). pCP25 complemented the four nonmotile mutants UW102-9, UW102-15, UW102-107, and UW102-146. The 4.5-kbp region was sequenced and four ORFs (gldA, orf1, orf2, and orf3) were identified (Fig. 2B). Further subcloning indicated that gldA was necessary for complementation of each mutant. pCP104 (which carried the BamHI/SalI fragment containing orf1 and half of gldA) and pCP105 (which contained the BamHI/XbaI fragment containing orfs 2 and 3 and the other half of gldA) did not complement any of the four nonmotile mutants described above (Fig. 2C). pSA11, which contained a 1.3-kbp fragment of DNA spanning the entire gldA gene, complemented UW102-9, UW102-15, UW102-107, and UW102-146 (Fig. 2C). The complemented mutants spread as well as wild-type colonies and exhibited rapid cell movements over glass or agar surfaces.
gldA Is Required for Gliding Motility.
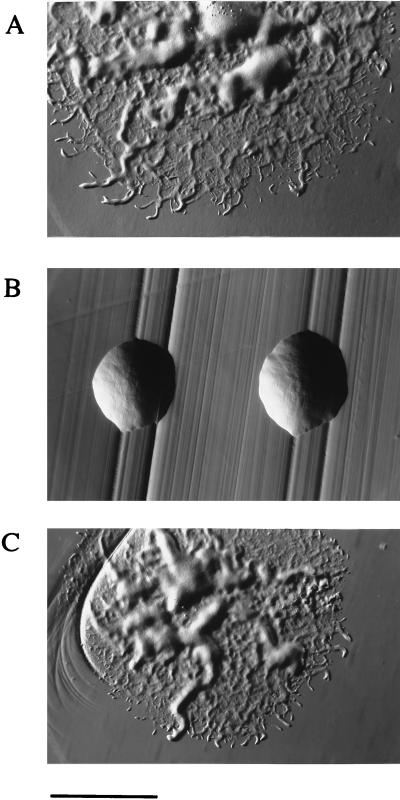
As described, we cloned an internal fragment of gldA into the suicide vector pLYL03 to generate pMM114. pMM114 was introduced into F. johnsoniae UW101 by conjugation and Em-resistant colonies arose after insertion into the chromosomal copy of gldA, generating the mutant CJ101-288. Disruption of gldA completely eliminated gliding motility, as measured by lack of colony spreading on PY2 agar (Fig. 3B) and by a complete lack of movement of individual cells or groups of cells over glass or agar surfaces. Motility was restored to CJ101-288 by pSA21, which carries the wild-type gldA gene (Fig. 3C).
Figure 3.
Photomicrographs of F. johnsoniae colonies. Colonies were grown for 2 days at 25°C on PY2 agar media. Photomicrographs were taken with an Olympus OM-4T camera mounted on a Nikon Diaphot inverted phase contrast microscope. (A) Wild-type F. johnsoniae UW101. (B) gldA knockout mutant CJ101-288. (C) CJ101-288 complemented with pSA21. (Bar = 1 mm.)
Isolation of the Tn4351-Induced gldA Mutant, CJ557.
In an independent attempt to identify gliding motility genes, we isolated nonmotile mutants after Tn4351 mutagenesis, by using procedures described (10). We isolated eight Tn4351 induced nonmotile mutants, cloned the disrupted genes in E. coli, and determined the nucleotide sequence of the regions near the transposon insertions (unpublished results). One of the mutants, CJ557, contained an insertion in gldA (Fig. 2B). The junction of the transposon facing the 3′ end of gldA was 11 nucleotides upstream of the predicted stop codon. Introduction of pSA21, which carries the wild-type gldA gene, into CJ557 restored motility as measured by colony spreading and by observations of single cell motility.
Phage Resistance of gldA Mutants.
It previously has been reported that many nonmotile mutants of F. johnsoniae are resistant to infection by a number of F. johnsoniae bacteriophage (23). The reason for this pleiotrophy is not known. We tested the sensitivity of F. johnsoniae strains UW101, UW102-09, CJ101-288, and CJ101-288 carrying pSA21, to the F. johnsoniae bacteriophage φCj1, φCj7, φCj13, φCj23, φCj28, φCj29, φCj42, φCj48, and φCj54. F. johnsoniae UW101 was readily lysed by these phage, whereas the nonmotile mutants UW102-9 and CJ101-288 were resistant to infection by each of them. Introduction of pSA21 into CJ101-288 restored sensitivity to each of these phage in addition to restoring gliding motility.
Movement of Latex Spheres by gldA Mutant Cells.
Wild-type cells of F. johnsoniae and related bacteria bind latex spheres on their surfaces and propel these spheres along the length of the cells (5, 24). We examined the ability of cells of the F. johnsoniae strains UW101, CJ101-288, and CJ101-288 carrying pSA21, to bind and propel latex spheres. Wild-type cells bound and propelled the spheres. Cells of the gldA mutant CJ101-288 were rarely observed to bind the spheres and were never observed to propel them. Cells of CJ101-288 carrying pSA21 bound and propelled the spheres as well as wild-type cells.
Analysis of the Sequence of gldA.
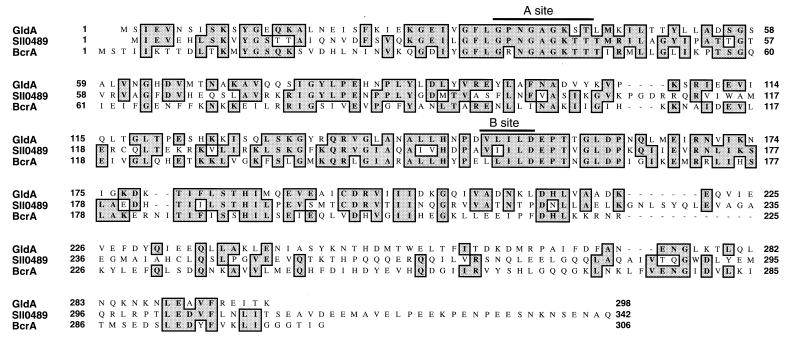
gldA is predicted to code for a 33.6-kDa protein containing 299 amino acid residues. Comparison of the predicted GldA protein to sequences in the databases revealed strong similarity to members of the ATP binding cassette family of transport proteins (ABC transport proteins) (Fig. 4, Table 2) (25–33). The region of strongest similarity spans the ATP-binding domains of these proteins (Walker Sites “A” and “B”), but the similarity to ABC transport proteins extends over the entire sequence of GldA. ABC transporters are found in bacterial, archael, and eukaryotic cells, where they transport a wide variety of molecules across one or more membranes either into or out of cells (25). The types of molecules that are transported by various ABC transporters include amino acids, peptides, proteins, polysaccharides, metals, and various drugs. Most bacterial ABC transporters are composed of several subunits that interact to form the final transport complex. GldA is similar to the ATP-binding components of ABC transporters, which are thought to provide the driving force for transport via ATP hydrolysis. The sequence that exhibits the greatest degree of similarity to GldA is the predicted protein product of orf sll0489 from the cyanobacterium Synechocystis PCC6803 (50% identity over 222 amino acid residues). This similarity is particularly interesting because Synechocystis PCC6803 exhibits the ability to move slowly over surfaces (34). The function of sll0489 is not known. Other proteins with known functions that exhibit high levels of similarity to GldA are listed in Table 2. Examination of the functions of these proteins did not allow us to make strong predictions regarding the exact function of GldA in gliding motility. GldA is probably involved in transport of some substance into or out of F. johnsoniae cells, but we do not know what this substance is. Analysis of the amino acid sequence of GldA revealed no obvious hydrophobic stretches, suggesting that GldA is not an integral membrane protein. GldA probably interacts with other transporter components that are anchored in the membrane. The ABC transporters listed in Table 2 are thought to have similar relationships with the transmembrane components of their multisubunit transport complexes.
Figure 4.
Alignment of the predicted amino acid sequence of GldA with sequences of putative ABC transport proteins. Amino acid residues that are identical are shaded. Sll0489 is the predicted protein product of Synechocystis PCC6803 orf sll0489. BcrA is the predicted sequence of the bcrA gene product from Bacillus licheniformis. Sites “A” and “B” are the “Walker A” and “Walker B” sites that are thought to be involved in ATP binding (25).
Table 2.
Comparison of GldA to various ABC transporters
| Protein | Organism | Similarity score | % amino acid identity with GldA | Function | Ref. |
|---|---|---|---|---|---|
| Sll0489 | Synechocystis PCC6803 | 694 | 50% identity over 222 residues | Unknown | 26 |
| BcrA | Bacillus licheniformis | 554 | 32.9% identity over 295 residues | Resistance to the peptide antibiotic bacitracin | 27 |
| MtrA | Streptomyces argillaceus | 496 | 34.2% identity over 260 residues | Resistance to the polyketide antibiotic mithramycin | 28 |
| NosF | Rhizobium meliloti | 478 | 38.4% identity over 216 residues | Cu processing/transport (for nitrous oxide reduction) | 29 |
| NodI | Bradyrhizobium japonicum | 475 | 34.8% identity over 221 residues | Oligosaccharide export (involved in nodulation) | 30 |
| NisF | Lactococcus lactis | 474 | 37% identity over 219 residues | Resistance to peptide antibiotic nisin | 31 |
| EpiF | Staphylococcus epidermidis | 422 | 34.6% identity over 205 residues | Resistance to the peptide antibiotic epidermin | 32 |
| CysA | Synechococcus PCC7942 | 412 | 33.5% identity over 212 residues | Uptake of sulfate | 33 |
Proteins were aligned with GldA and the similarity scores were determined using the lalign program (22).
Analysis of the Region of DNA Surrounding gldA.
The genes for the individual components of bacterial ABC transporters often are clustered together on the chromosome in one or more operons (25). For this reason, we examined the region of DNA surrounding gldA. Two ORFs (orf2 and orf3) lie upstream of gldA (Fig. 2B). These ORFs are oriented in the opposite direction to gldA and are separated from gldA by 148 bp. The predicted protein products of orfs 2 and 3 are 194 and 193 amino acid residues long, respectively and are very similar to each other (66% identical over their entire sequences). The predicted proteins Orf2 and Orf3 share weak but significant sequence similarity (24.7% and 20.2% sequence identity, respectively) with the Ail protein of Yersinia enterocolitica (35). Ail is an outer membrane protein that is thought to be involved in adhesion of Y. enterocolitica to mammalian cells. We do not have any evidence that orf2 or orf3 are involved in F. johnsoniae gliding motility.
There are no obvious ORFs immediately downstream of gldA. A perfect 15-nucleotide inverted repeat sequence that is followed by a string of T’s (AACCCAATTTATTCCTGGAATAAATTGGGTTTTCTTTTTT) is found 38 nucleotides downstream from the gldA stop codon. This may function as a transcription terminator. We identified a large ORF (orf1)that started 388 bp downstream of the gldA stop codon and was oriented in the same direction. Partial sequence of orf1 suggests that it codes for a protein containing greater than 579 amino acid residues. The predicted protein has weak but significant similarity (20% amino acid identity over 396 amino acid residues) to the IutA protein of E. coli (36). IutA is an outer membrane protein that is thought to interact with TonB and play a role in transport of iron into the cell. Because orf1 may code for a protein involved in transport and lies downstream of gldA, it seemed possible that the orf1 gene product might interact with GldA and be required for gliding motility. To test this possibility we disrupted the chromosomal copy of orf1 in F. johnsoniae UW101 as described in Materials and Methods. Disruption of orf1 had no obvious effect on gliding motility (data not shown). Colonies spread as well as those of the wild type, and individual cells exhibited rapid motility over glass and agar surfaces.
Possible Functions of gldA in Gliding Motility.
The results presented above identify one gene, gldA, that is essential for gliding motility in F. johnsoniae. There are many possible roles for GldA. (i) GldA may export polysaccharide. Exopolysaccharides and cell surface polysaccharides have been implicated in gliding motility (3, 7, 37). They may be essential in allowing the cell to adhere to or move along a surface. (ii) GldA could function to export protein components of the gliding machinery to the periplasm or cell surface. (iii) GldA could be a functional part of the gliding “motor” that provides the thrust to move the cell. It is unlikely, however, that the predicted ATPase activity of GldA drives cell movement because the available evidence indicates that proton motive force, rather than ATP hydrolysis, is the driving force for F. johnsoniae gliding motility (5). Given the wide range of functions of ABC transporters in bacteria and the many models that have been proposed to explain gliding motility, many other possible roles for GldA exist. The analysis of other genes involved in F. johnsoniae gliding motility should help determine the exact function of gldA. Our current model of F. johnsoniae gliding motility, based on observations of cell movements, is similar to the one proposed by Lapidus and Berg (24). It involves the movement of adhesion sites in the outer membrane along tracks that are anchored to the cell wall. Additional proteins in the cytoplasmic membrane and periplasm are postulated to harness the proton motive force and perform the work that is necessary to move the adhesion sites.
Gliding bacteria are found in many of the branches of the eubacterial phylogenetic tree. Genetic analysis of gliding motility has until now been confined to Myxococcus xanthus, which is not closely related to F. johnsoniae. M. xanthus appears to have two independent motility systems (“A” and “S”) that allow cells to move under different conditions (38, 39). “S” motility requires polar pili and may be related to another form of surface translocation called twitching motility (40). “A” motility does not require pili. The exact mechanism responsible for cell movement is not known for either M. xanthus motility system. The situation may be somewhat simpler in F. johnsoniae. Mutations in single genes of F. johnsoniae eliminate gliding motility, which may suggest that F. johnsoniae has a single motility system. The relationship of the F. johnsoniae motility system to either of the M. xanthus systems is not known.
The results reported in this paper describe the identification of a gene required for F. johnsoniae gliding motility. gldA complements 4 of 61 independently isolated nonmotile mutants. We recently identified another gene, gldB, that complements four different nonmotile mutants (D.W.H. and M.J.M., unpublished data). These results may indicate that there are a limited number of genes (perhaps less than 20) required for gliding motility. Continued analysis of these genes and their gene products should help to elucidate the mechanism of F. johnsoniae gliding motility.
Acknowledgments
This research was supported by a grant from the National Science Foundation (MCB-9418308) and by a Shaw Scientist Award to M.J.M. from the Milwaukee Foundation. Some of the DNA sequencing was performed by the Automated DNA Sequencing Facility of the University of Wisconsin, Milwaukee, Department of Biological Sciences.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: Em, erythromycin; ABC, ATP binding cassette.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF007381).
References
- 1.Reichenbach H, Ludwig W, Stackebrandt E. Arch Microbiol. 1986;145:391–395. [Google Scholar]
- 2.Burchard R P. Annu Rev Microbiol. 1981;35:497–529. doi: 10.1146/annurev.mi.35.100181.002433. [DOI] [PubMed] [Google Scholar]
- 3.Pate J L. Can J Microbiol. 1988;34:459–465. [Google Scholar]
- 4.Bernardet J-F, Segers P, Vancanneyt M, Berthe F, Kersters K, Vandamme P. Int J Syst Bacteriol. 1996;46:128–148. doi: 10.1099/00207713-46-3-782. [DOI] [PubMed] [Google Scholar]
- 5.Pate J L, Chang L-Y E. Curr Microbiol. 1979;2:59–64. [Google Scholar]
- 6.Abbanat D R, Leadbetter E R, Godchaux W, III, Escher A. Nature (London) 1986;324:367–369. [Google Scholar]
- 7.Godchaux W, III, Gorski L, Leadbetter E R. J Bacteriol. 1990;172:1250–1255. doi: 10.1128/jb.172.3.1250-1255.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang L Y E, Pate J L, Betzig R J. J Bacteriol. 1984;159:26–35. doi: 10.1128/jb.159.1.26-35.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wolkin R H, Pate J L. J Gen Microbiol. 1985;131:737–750. [Google Scholar]
- 10.McBride M J, Kempf M J. J Bacteriol. 1996;178:583–590. doi: 10.1128/jb.178.3.583-590.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McBride M J, Baker S A. Appl Environ Microbiol. 1996;62:3017–3022. doi: 10.1128/aem.62.8.3017-3022.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pate J L, Petzold S J, Chang L-Y E. Curr Microbiol. 1979;2:257–262. [Google Scholar]
- 13.Bolivar F, Backman K. Methods Enzymol. 1979;68:245–267. doi: 10.1016/0076-6879(79)68018-7. [DOI] [PubMed] [Google Scholar]
- 14.Simon R, Priefer U, Puhler A. Bio/Technology. 1983;2:784–791. [Google Scholar]
- 15.Vieria J, Messing J. Gene. 1982;19:259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- 16.Hedges R W. Mol Gen Genet. 1974;132:31–40. doi: 10.1007/BF00268228. [DOI] [PubMed] [Google Scholar]
- 17.Li L-Y, Shoemaker N B, Salyers A A. J Bacteriol. 1995;177:4992–4999. doi: 10.1128/jb.177.17.4992-4999.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reeves A R, D’Elia J N, Frias J, Salyers A A. J Bacteriol. 1996;178:823–830. doi: 10.1128/jb.178.3.823-830.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 20.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 21.Pearson W R. Methods Enzymol. 1990;183:63–98. doi: 10.1016/0076-6879(90)83007-v. [DOI] [PubMed] [Google Scholar]
- 22.Huang X, Miller W. Adv Appl Math. 1991;12:337–357. [Google Scholar]
- 23.Wolkin R H, Pate J L. J Gen Microbiol. 1986;132:355–367. [Google Scholar]
- 24.Lapidus I R, Berg H C. J Bacteriol. 1982;151:384–398. doi: 10.1128/jb.151.1.384-398.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fath M J, Kolter R. Microbiol Rev. 1993;57:995–1017. doi: 10.1128/mr.57.4.995-1017.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaneko T. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 27.Podlesek Z, Comino A, Herzog-Velikonja B, Zgur-Bertok D, Komel R, Grabnar M. Mol Microbiol. 1995;16:969–976. doi: 10.1111/j.1365-2958.1995.tb02322.x. [DOI] [PubMed] [Google Scholar]
- 28.Fernendez E, Lombe F, Mendez C, Salas J A. Mol Gen Genet. 1996;251:692–698. doi: 10.1007/BF02174118. [DOI] [PubMed] [Google Scholar]
- 29.Holloway P, McCormick W, Watson R J, Chan Y K. J Bacteriol. 1996;178:1505–1514. doi: 10.1128/jb.178.6.1505-1514.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gottfert M, Hitz S, Hennecke H. Mol Plant–Microbe Interact. 1990;3:308–316. doi: 10.1094/mpmi-3-308. [DOI] [PubMed] [Google Scholar]
- 31.Siegers K, Entian K D. Appl Environ Microbiol. 1995;61:1082–1089. doi: 10.1128/aem.61.3.1082-1089.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peschel A, Gotz F. J Bacteriol. 1996;178:531–536. doi: 10.1128/jb.178.2.531-536.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Green L S, Laudenbach D E, Grossman A R. Proc Natl Acad Sci USA. 1989;86:1949–1953. doi: 10.1073/pnas.86.6.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stanier R Y, Kunisawa R, Mandel M, Cohen-Bazire G. Bacteriol Rev. 1971;35:171–205. doi: 10.1128/br.35.2.171-205.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller V L, Bliska J B, Falkow S. J Bacteriol. 1990;172:1062–1069. doi: 10.1128/jb.172.2.1062-1069.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krone W J A, Stegehuis F, Koningstein G, Van Doorn C, Roosendaal B, De Graaf F K, Oudega B. FEMS Microbiol Lett. 1985;26:153–161. [Google Scholar]
- 37.Godchaux W, III, Lynes M A, Leadbetter E R. J Bacteriol. 1991;173:7607–7614. doi: 10.1128/jb.173.23.7607-7614.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hodgkin J, Kaiser D. Mol Gen Genet. 1979;171:177–191. [Google Scholar]
- 39.Shi W, Zusman D R. Proc Natl Acad Sci USA. 1993;90:3378–3382. doi: 10.1073/pnas.90.8.3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu S S, Kaiser D. Mol Microbiol. 1995;18:547–558. doi: 10.1111/j.1365-2958.1995.mmi_18030547.x. [DOI] [PubMed] [Google Scholar]