Abstract
Our laboratory has previously reported that UVA irradiation can increase the expression of Bcl-XL, an antiapoptotic molecule, by stabilizing its mRNA in cultured immortalized human keratinocytes. To understand the mechanism by which the Bcl-XL message is stabilized, we used a synthetic Bcl-XL 3′-untranslated region (UTR) to capture RNA-binding proteins. Nucleolin was identified as one of the binding proteins as determined by tandem mass spectrometry coupled to liquid chromatography analysis. Further study showed that nucleolin specifically recognized the AU-rich elements (AUUUA) in the 3′-UTR of the Bcl-XL mRNA and could stabilize the mRNA in vitro. Furthermore, overexpression of nucleolin stabilizes the Bcl-XL mRNA in HeLa cells, whereas reducing nucleolin by small interfering RNA shortens the Bcl-XL mRNA half-life. Interestingly, nucleolin physically interacted with polyadenylate [poly(A)]–binding protein through it RGG motifs. Its stabilizing effect on the Bcl-XL mRNA was dependent upon the presence of poly(A) tail. Based on these data, we propose a model in which nucleolin protects the Bcl-XL mRNA from nuclease degradation by enhancing the stability of the ribonucleoprotein loop structure.
Introduction
Skin cancer has become a serious health problem in the United States. Each year, there are more than one million new cases of skin cancer diagnosed, accounting for 40% of all cancer cases. Solid evidence has shown that UV irradiation from sunlight is the primary carcinogen for skin cancer (1–3). The UV irradiation can be categorized into UVA (320−400 nm), UVB (280−320 nm), and UVC (200−280 nm), based on its wavelength. UVA, composed of the vast majority of the irradiation from sunlight (90−99%), has been shown to be a potent skin carcinogen (3–5). For example, UVA promotes malignant transformation in cultured human keratinocytes (HaCaT cells; ref. 6) and causes malignant melanoma and squamous cell carcinoma in mouse models (7, 8). Specifically, UVA causes DNA damage by increasing reactive oxygen species and producing cyclobutane pyrimidine dimers (4, 9, 10). Additionally, UVA activates multiple signaling pathways, i.e., phosphoinositide 3-kinase, p38, and c-Jun-NH2-kinase (JNK), important for cell survival upon UVA irradiation (6, 11, 12).
The antiapoptotic molecule Bcl-XL is crucial for the survival of many types of cells and has been implicated in differentiation and development (13, 14). For example, knockout of Bcl-XL is lethal in mice, resulting from extensive death of hematopoietic cells and atrophy of the brain (15). On the other hand, substantial induction of this molecule renders activated T cells resistant to apoptosis upon CD28 stimulation (16). Its importance can be further illustrated by its involvement in cancer development. Overexpression of Bcl-XL is observed in several types of cancers, i.e., colorectal and breast cancer (14, 17). The importance of Bcl-XL in skin carcinogenesis has been well defined in both cultured cells and animal models (18, 19). Furthermore, Bcl-XL confers drug resistance in multiple cancers (20, 21) and inversely correlates with prognosis in some cancers (22). Therefore, a thorough understanding of the regulation of Bcl-XL will pave the way for novel strategies of cancer chemotherapy and chemoprevention.
Bcl-XL primarily localizes to the mitochondrial membrane. Through its BH1−3 domains, Bcl-XL is able to bind and sequester proapoptotic molecules possessing the BH-3 domain (23). The primary targets for Bcl-XL are Bax and Bak, which migrate to and oligomerize on the outer mitochondrial membrane and thus change the permeability of the mitochondria, leading to the release of small molecules, including cytochrome c. The release of cytochrome c triggers the assembly of apoptosomes and, thus, activation of caspase cascade (13, 14, 17). It has been postulated that Bcl-XL blocks the oligomerization of Bax and Bak and, thus, the release cytochrome c (23).
The expression of Bcl-XL is tightly regulated at transcriptional (24, 25), alternatively splicing (24), and translational levels (16). Recently, our laboratory has shown that its mRNA stability can also be regulated in human keratinocytes upon irradiation with 250 kJ/m2 UVA. Furthermore, this stabilization is dependent upon the 3′-UTR of the Bcl-XL mRNA (26). However, the mechanism for the mRNA stabilization of the Bcl-XL mRNA is unclear.
The regulation of mRNA stability enables cells to rapidly adjust to environmental changes (27, 28). Indeed, mRNAs of some regulatory molecules, such as c-myc, cyclins, p27, cyclooxygenase-2 (Cox-2), and interleukin 2 (IL-2), are normally short-lived, and their stability is subject to change upon external stimulation (29). In mammalian cells, the rate-limiting step of mRNA degradation is polyadenylate [poly(A)] deadenylation, which is mediated by poly(A) RNase (PARN; ref. 28). Shortening of poly(A) tail to ∼30 to 60 nucleotides in mammalian cells is required for mRNA degradation (30). After deadenylation, hydrolysis of 5′ m7G cap takes place, allowing degradation of decapped mRNA by 5′-3′ exoribonuclease, Xrn1 (31–33). However, it has been argued that the primary degradation pathway in mammalian cells is mediated by exosomes, complexes composed of at least ten 3′-5′ exonucleases (28, 31, 32).
It has been shown that stability of many mRNAs is dependent upon their 3′-UTRs, including those of cyclins, Cox-2, IL-2, renin, c-myc, IL-6, granulocyte macrophage colony–stimulating factor, ferritin, and glucose transport 1 (27–29, 34). Prominently, a cis-element, AU-rich element (ARE), has been well defined and shown to be critical for the stability of these mRNAs. It was estimated that 5% to 8% of all mRNAs contain variable numbers of AREs (27, 34).
Despite the fact that ARE was identified >15 years ago, the mechanism of how it functions was not unraveled until recently. Several studies related to known ARE-binding proteins indicated that some ARE-binding proteins physically interact and thus recruit RNA decay machinery to accelerate RNA degradation. For example, K homology splicing regulatory protein (KSRP) has been known as a destabilizing factor. In a recent study, Chou et al. made a fusion protein between KSRP and MS2, a bacteriophage coat protein. This fusion protein led to a dramatic decrease in the β-globin message that contained six MS2 binding sites. Furthermore, they found that PARN and RRP4 exosome components could be coimmunoprecipitated with KSRP (35). These data suggested that KSRP accelerates mRNA degradation by recruiting the PARN and exosomes. Similarly, other studies showed that two other destabilizing ARE-binding proteins, Tristetraprolin and ARE/poly(U)-binding/degradation factor 1 (AUF1), could also associate with exosomes (31).
Meanwhile, a number of studies also showed a stabilizing function of AREs (36–39). For example, HuR can stabilize a number of messages, such as β-casein, SLC11A1, p21, cyclins A and B1, Cox-2, and renin, through the ARE element (40). One ubiquitous protein, nucleolin, has also been reported to stabilize several messages, such as Bcl-2, IL-2, and Gadd45 (37, 38, 41).
To understand how the Bcl-XL mRNA was stabilized upon UVA irradiation in human keratinocytes (HaCaT cells), we identified nucleolin as one of the proteins binding to the 3′-UTR of the mRNA. In this report, we provide evidence that nucleolin can bind to the ARE element on the 3′-UTR of the Bcl-XL mRNA and stabilize the message in vitro and in vivo. Moreover, nucleolin stabilization of the Bcl-XL mRNA is dependent on the poly(A) tail and poly(A) binding protein (PABP). Finally, we offer a model to explain how nucleolin stabilizes the Bcl-XL mRNA.
Materials and Methods
Plasmids
Constructs expressing the wild-type and ARE-mutated Bcl-XL (NM_001191) 3′-UTR (1287nt) were cloned as follows. The UTRs were dropped out by EagI from the UTR-luciferase reporter constructs (26) and then religated to NotI-digested pBluescript II KS (—) plasmid (Stratagene). The construct was named pBS-1287UTR(WT). The full length Bcl-XL was cloned by reverse transcription–PCR (RT-PCR). Briefly, total RNA extracted from HaCaT cells (immortalized human keratinocytes) with Trizol (Invitrogen) was reverse-transcribed with ProtoScript II RT-PCR kit (New England Biolabs). The primers for the subsequent PCR amplification were 5′-catgcggcgcggaggaggaagcaagcgagg-3′ and 5′-ctctcgagcactgagtaaacacagtttattactg-3′ to amplify the full-length mRNA with Deep Vent DNA polymerase (New England Biolabs). The PCR product (3542nt) was then cloned into pCR4-TOPO vector (Invitrogen) to make the construct TOPO-Full(WT). The pEGFP-nucleolin vector for transfection was a generous gift from Dr. Kastan of St. Jude's Children's Hospital. Recombinant nucleolin was expressed from MBP-Nucleolin-pMalC2 vector, a generous gift from Dr. Maizels of the University of Washington and purified with amylase resin (New England Biolabs; refs. 42, 43). The recombinant His6-PABP was expressed from the plasmid pET28a-PABP, kindly provided by Dr. Kiledjian of Rutgers University (44), and purified with Ni-NTA agarose (Qiagen).
Mutagenesis
The three AREs on both the constructs pBS-1287UTR(WT) and TOPO-Full(WT) were mutated with Quickchange kit (Stratagene). The primer sets to mutate the AREs were 5′-cttgcagctagttttctagaacccatcacacttctgtgagaccc-3′ and 5′-gggtctcacagaagtgtgatgggttctaaaactagctgcaag-3′ (ARE1), 5′-ccccctaagagccacccaggggccactttttgact-3′ and 5′-agtcaaaagtggcccctgggtggctcttaggggg (ARE2), and 5′-ttttgttaagcgtgtctgtacccatgtgtgaggagctgctgg-3′ and 5′-cagcagctcctcacacatgggtacagacacgcttaacaaaa-3′ (ARE3).
RNA affinity chromatography
UVA and SB202190 treatment of HaCaT cells (immortalized human keratinocytes) was described previously (26). These samples were subject to the RNA affinity chromatography as described by Cok et al. (45). Briefly, the treated cells were washed twice with cold PBS and scraped off the plate with the EMSA binding buffer [10 mmol/L HEPES (pH 7.6), 5 mmol/L MgCl2, 40 mmol/L KCl, 1 mmol/L DTT, 5% glycerol, 5 mg/mL heparin]. The cells were sonicated and centrifuged. The supernatant was used for the subsequent RNA binding assay. To transcribe Bcl-XL 3′-UTR in vitro, the construct pBS-1287UTR(WT) was completely digested by XhoI and phenol extracted. This linearized DNA template was transcribed and biotinylated with MEGAscript T7 kit (Ambion) according to the manufacturer's instructions. Subsequently, 500 μg cell lysate were incubated with 20 μg of biotinated RNA on ice for 1 h. The mixture was further incubated with 0.2 mL Streptavidin Sepharose beads (Amersham) overnight at 4°C with rotation. The beads were loaded on a column and washed with 20 mL of the binding buffer. The beads were boiled with SDS-PAGE loading buffer to release captured proteins, which were resolved on a 10% SDS-PAGE gel. The proteins were silver-stained with a mass spec compatible kit (Invitrogen) and those differentially captured bands were excised for mass spec analysis.
Tandem mass spectrometry coupled to liquid chromatography
Excised protein bands after SDS-PAGE were digested in trypsin (10 μg/mL) at 37°C overnight. Tandem mass spectrometry coupled to liquid chromatography (LC-MS/MS) analyses of in-gel trypsin digestion was performed as previously described (46). Protein bands were analyzed using a quadrupole ion trap ThermoFinnigan LCQ DECA XP PLUS equipped with a Michrom Paradigm MS4 HPLC and a nanoelectrospray source. Peptides were eluted from a 15-cm pulled tip capillary column (100 μm I.D. × 360 μm O.D.; 3−5 μm tip opening) packed with 7 cm Vydac C18 material (5 μm, 300 Å pore size), using a gradient of 0 to 65% solvent B (98% methanol:2% water:0.5% formic acid:0.01% triflouroacetic acid) over a 60-min period at a flow rate of 350 nL/min. The LCQ DECA XP PLUS electrospray positive mode spray voltage was set at 1.6 kV, and the capillary temperature was at 200°C. Dependent data scanning was performed by the Xcalibur v 1.3 software (47) with a default charge of 2, an isolation width of 1.5 amu, an activation amplitude of 35%, an activation time of 30 ms, and a minimal signal of 10,000 ion counts. Global dependent data settings were as follows: reject mass width of 1.5 amu, dynamic exclusion enabled, exclusion mass width of 1.5 amu, repeat count of 1, repeat duration of 1 min, and exclusion duration of 5 min. Scan event series included one full scan with mass range of 350 to 2,000 Da, followed by three dependent MS/MS scan of the most intense ion. Tandem MS spectra of peptides were analyzed with Turbo SEQUEST v 3.1, a program that allows the correlation of experimental tandem MS data with theoretical spectra generated from known protein sequences (48). The peak list (data files) for the search was generated by Xcalibur 1.3. Parent peptide mass error tolerance was set at 1.5 amu, and fragment ion mass tolerance was set at 0.5 amu during the search. The criteria that were used for a preliminary positive peptide identification are the same as previously described, namely peptide precursor ions with a +1 charge having an Xcorr of >1.8, +2 Xcorr of > 2.5, and +3 Xcorr of >3.5. A dCn score of >0.08 and a fragment ion ratio of experimental/theoretical of >50% were also used as filtering criteria for reliable matched peptide identification (49). All matched peptides were confirmed by visual examination of the spectra. All spectra were searched against the latest version of the nonredundant protein database downloaded on July 14, 2005 from National Center for Biotechnology Information (NCBI). At the time of the search, the nonredundant protein database from NCBI contained 2,662,317 entries.
UV-crosslinking assay
The Bcl-XL 3′-UTR was synthesized and 32P-labeled from XhoI linearized pBS-1287(WT) plasmid with the Riboprobe In vitro Transcription System (Promega). After removal of unincorporated nucleotides by P-6 minicolumn (Bio-Rad), 5 million counts per million were incubated with 0.1 μg of MBP-Nucleolin on ice for 30 min, followed by UV cross-linking by Genelinker (Bio-Rad). RNase A1 (2.5 μg; Sigma) was added to the reaction and incubated at 37°C for 1 h. The reaction was resolved on a SDS-PAGE gel and autoradiographed.
RNA immunoprecipitation and RT-PCR
RNA immunoprecipitation was performed as previously described (41). Briefly, HaCaT cells were lysed with radioimmunoprecipitation assay (RIPA) buffer with protease inhibitors. Antibodies (2 μg) were incubated with 500 μg lysates for 4 h at 4°C. Protein A/G beads (Santa Cruz Biotech) were added and incubated overnight at 4°C. The beads were washed five times with RIPA buffer, and RNA was extracted with Trizol reagent (Invitrogen). The extracted RNA was reverse-transcribed with the Protoscript First Strand cDNA synthesis kit (New England Biolabs) and followed by PCR to generate a Bcl-XL specific fragment with the primer set: cattgccaccaggagaaccactac (forward) and catctttatcccaagcagcct (reverse).
Cell transfection
The transfection of pEGFP-nucleolin (generous gift from Dr. Kastan) was performed with LipofectAMINE 2000 (Invitrogen) according to the manufacturer's instruction. The transfection of the nucleolin and control small interfering RNA (siRNA; Qiagen) was performed with HiPerFect (Qiagen) based on the manufacturer's protocol. The siRNA effects were monitored 60 h after transfection by Western blot analysis.
RT-PCR and real-time PCR
The RNA was extracted with Trizol reagent (Invitrogen) and reverse-transcribed with high-capacity cDNA reverse transcription kit (Applied Biosystems, Inc.) according to the manufacturers' instructions. The real-time PCR was performed with Taqman probes for human Bcl-XL and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) on the ABI PRISM 7700 sequence detection system (Applied Biosystems). The mRNA level of Bcl-XL was normalized to GAPDH in each sample according to the User Bulletin 2 of ABI PRISM 7700 sequence detection system.
Western blot analysis and immunoprecipitation
The Western blot analysis and immunoprecipitation were described previously (26). The monoclonal antibodies against nucleolin (MS-3, D6) and PABP (10E10), as well as protein A/G beads, were purchased from Santa Cruz Biotech.
In vitro RNA degradation assays
The experiments were performed as described by Chen et al. (41). Briefly, the full-length Bcl-XL mRNA (∼ 3500nt) was transcribed in vitro from the XbaI-linearized TOPO-Full(WT) with mMessage mMachine T7 kit (Ambion). Next, 10 million counts per minute of the 32P-labeled Bcl-XL mRNA (5′-capped and 3′-polyadenylated) were incubated at 37°C with 20 μg of S100 fraction of HaCaT cells in 25 μL of EMSA buffer in the presence or absence of 5 μg MBP-nucleolin. Samples were taken at the indicated times and resolved on SDS-PAGE gel, followed by autoradiography.
Results
Nucleolin is identified as one of the proteins binding to the Bcl-XL 3′-UTR
Our laboratory has reported that in HaCaT cells, Bcl-XL mRNA is stabilized through its 3′-UTR upon UVA irradiation (26). Because 3′-UTRs confer mRNA stability by interacting with different RNA-binding proteins (27, 34, 50), we hypothesized that the Bcl-XL mRNA was stabilized by a similar mechanism. Specifically, there might be more stabilizing factor binding to the Bcl-XL 3′-UTR upon UVA irradiation in HaCaT cells. To test this idea, we started with identifying RNA-binding proteins that could be differentially captured from the mock-treated or UVA-treated HaCaT cells. Moreover, because this stabilization was dependent upon p38 mitogen-activated protein kinase (MAPK; ref. 26), we inhibited p38 MAPK with 5 μmol/L of SB202190 in the UVA-treated cells. We used a synthetic 1287nt wild-type Bcl-XL 3′-UTR to capture RNA-binding proteins from the above cell lysates. After the proteins were resolved by SDS-PAGE and silver-stained, we found that there were at least 10 protein bands that were differentially captured (Fig. 1, lanes 1 and 2). One of the captured proteins increased by UVA irradiation and inhibited by SB202190 was excised and identified with LC-MS/MS.
Figure 1.
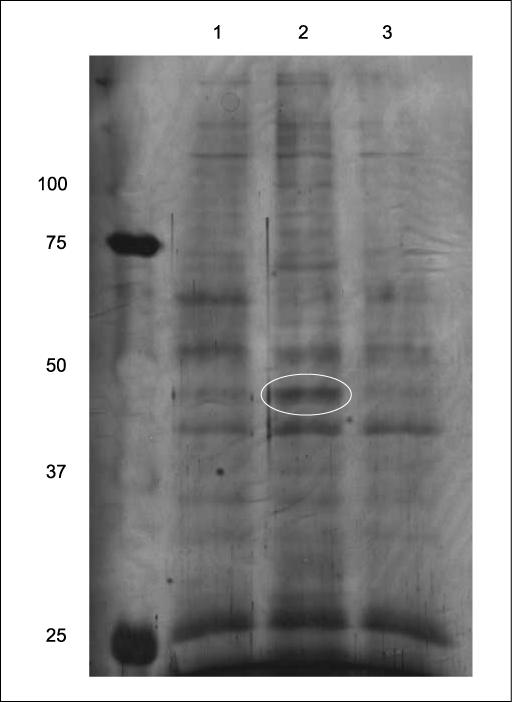
Nucleolin was identified as one of the proteins binding to the 3′-UTR of Bcl-XL mRNA using RNA affinity chromatography. HaCaT cells were either mock-irradiated or UVA-irradiated (250 kJ/m2), in the presence of DMSO (lanes 1 and 2) or 5 μmol/L of SB202190 (lane 3). The cells were lysed 2 h postirradiation. Proteins (500 μg) were incubated with 5 μg of the biotinylated Bcl-XL 3′-UTR for 1 h on ice and then with 0.2 mL of Streptavidin agarose beads for 4 h at 4°C. The beads were loaded on a column and washed with 10 mL of the EMSA buffer before boiling with SDS-PAGE sample buffer to release binding proteins that were silver stained after being resolved with a 10% gel. Lane 1, mock-irradiated sample; lane 2, UVA-irradiated sample; lane 3, UVA-irradiated sample plus SB202190. The bands that were differentially stained were excised from the gel for the subsequent mass spectrometry analysis. The encircled band was identified as nucleolin.
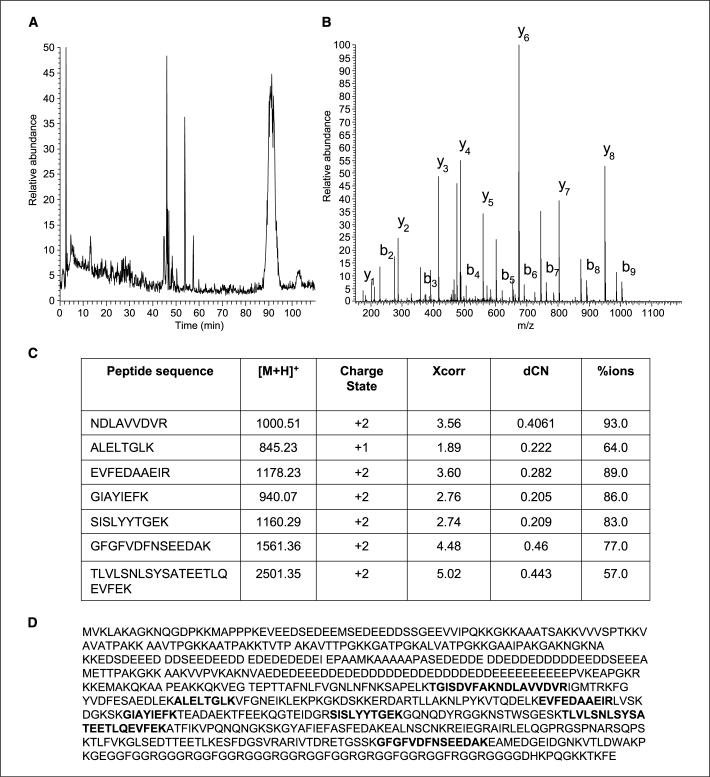
The encircled band in Fig. 1 was subsequently identified as nucleolin. Figure 2A showed the LC-MS/MS trace of the trypsin-digested SDS-PAGE protein band. A search of the MS/MS spectra (see Materials and Methods) identified seven peptides from nucleolin, mapping 14% of the primary protein sequence (Fig. 2B and C). The MS/MS spectra of these peptides showing the various fragment ions that were found are shown in Fig. 2D.
Figure 2.
Mass spectrometry base peak chromatogram (LC-MS) of the in-gel digested SDS-PAGE band corresponding to nucleolin. A, a typical MS/MS spectrum for one of the peptides identified from nucleolin is also shown along with its corresponding b/y fragment ions used in determining the sequence (B). A total of seven high-scoring peptides (C) as described in Materials and Methods were identified covering 14% (bold) of the primary sequence of nucleolin (D).
To validate the mass spectrometry results, we used Western blot analysis to detect the presence of nucleolin from the RNA affinity chromatography. As expected, we found that the specific anti-nucleolin antibody recognized nucleolin (Fig. 3A, top). Furthermore, we noticed that nucleolin captured from the UVA-treated sample was more abundant than that from the mock-treated sample (Fig. 3A, lane 3, top), given that the input nucleolin was equal (Fig. 3A, bottom). These data indicated that UVA treatment enhanced the nucleolin binding to the 3′-UTR of the Bcl-XL mRNA and thus supported our initial hypothesis. This is consistent with the previous finding that UV increases the nucleolin binding to TAR (HIV-1 transactivation responsive region) RNA probe in RKO cells (51).
Figure 3.
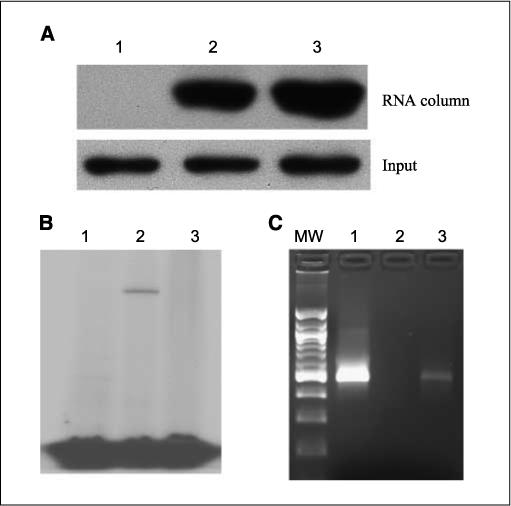
Nucleolin binds to the Bcl-XL 3′-UTR in vitro and in vivo. A, HaCaT cells were harvested 2 h after mock or UVA irradiation (250 kJ/m2). Cell lysates (100 μg) were incubated with (lanes 2 and 3) or without (lane 1) 1 μg of the biotinated Bcl-XL 3′-UTR for 1 h on ice and 50 μL of Streptavidin beads for 4 h at 4°C. The beads were washed extensively, and the captured proteins were examined by Western blot analysis (top). Lane 2, mock treatment; lane 3, UVA irradiation. B, recombinant nucleolin (1 μg) was incubated with 32P-labeled 3′-UTR before UV cross-linking. The RNase A was added to the reaction and incubated for 30 min. The reaction was resolved on SDS-PAGE gel and subject to autoradiography. Lanes 1 and 2, the wild type 3′-UTR; lane 3, mutated 3′-UTR (3× ARE). Lane 1, no UV cross-linking; lane 2, UV cross-linking of the wild-type 3′-UTR; lane 3, UV cross-linking of ARE mutated 3′-UTR. C, RNA immunoprecipitation followed by RT-PCR. Endogenous nucleolin was pulled down with the antinucleolin antibody and protein A/G beads. The RNA was extracted from the beads and reverse-transcribed into cDNA. PCR was performed to imply a 500-bp fragment with specific Bcl-XL primers.
To rule out the possibility that RNA affinity chromatography might capture nonspecific proteins, we used the UV cross-linking assay to prove the specific binding of nucleolin to the 3′-UTR of Bcl-XL. The cross-linking between protein and RNA would form covalent bonds between protein and RNA and thus shield the protein-bound RNA region from subsequent nuclease A1 digestion (52). As shown in Fig. 3B, when there was no UV cross-linking, the RNA was completely degraded (Fig. 3B, lane 1). In contrast, after being cross-linked, a band appeared at 90 to 100 kDa, the predicted size of the recombinant nucleolin, indicating that the recombinant nucleolin binds to the 3′-UTR and therefore protected its binding region from nuclease digestion.
Nucleolin binds to AREs of the 3′-UTR
Next, we sought to investigate what cis-element on the 3′-UTR nucleolin bound. It has been reported that nucleolin binds to ARE of several other mRNA molecules, including Bcl-2 (37, 38). We found that there are three AUUUA elements on the 3′-UTR of Bcl-XL. To test if nucleolin also binds to these ARE elements, we mutated all of the three AREs from AUUUA to ACCCA and used the mutated version for the protection assay. We found that nucleolin failed to protect any region when all of the three AREs were mutated (Fig. 3B, lane 3). Therefore, the result clearly showed that the recombinant nucleolin could bind to the AREs of the Bcl-XL 3′-UTR in vitro. Moreover, nucleolin could still protect RNA with single or double ARE mutations (data not shown), suggesting that nucleolin binding to single ARE could protect the RNA from degradation.
Nucleolin binds to the Bcl-XL 3′-UTR in vivo
The above result was achieved in vitro with the recombinant protein. We next asked the question if the endogenous nucleolin could also bind the Bcl-XL 3′-UTR in intact cells. To answer this question, we used the RNA immunoprecipitation to pull down nucleolin together with its potential mRNA targets. This technique has been successfully used to examine in vivo RNA-protein interactions (53, 54). After RT-PCR amplification with the Bcl-XL specific primers, we found that the sample immunoprecipitated with the antinucleolin antibody generated a band of the expected size (∼ 500 bp), whereas the sample immunoprecipitated with the mouse IgG failed to yield the specific band (Fig. 3C). This result showed that Bcl-XL mRNA was bound by endogenous nucleolin in intact cells.
Nucleolin can stabilize Bcl-XL mRNA RNA both in vitro and in vivo
We wanted to understand the biological significance of the nucleolin binding to the 3′-UTR of Bcl-XL message. Nucleolin has been shown to stabilize mRNAs of several molecules, including Bcl-2 (37, 38, 41). Therefore, it was possible that nucleolin could also stabilize the Bcl-XL mRNA. To test this hypothesis, we transcribed the full-length Bcl-XL (3500nt, including 5′-UTR, coding region, and 3′-UTR) in vitro. This m7G capped and polyadenylated RNA was incubated with S100 cellular fraction in the presence or absence of recombinant nucleolin. As shown in Fig. 4A, the half-life of the Bcl-XL mRNA increased from ∼2 h (without nucleolin) to ∼4 h (with nucleolin). Therefore, nucleolin was able to stabilize the Bcl-XL mRNA in vitro (Fig. 4A).
Figure 4.
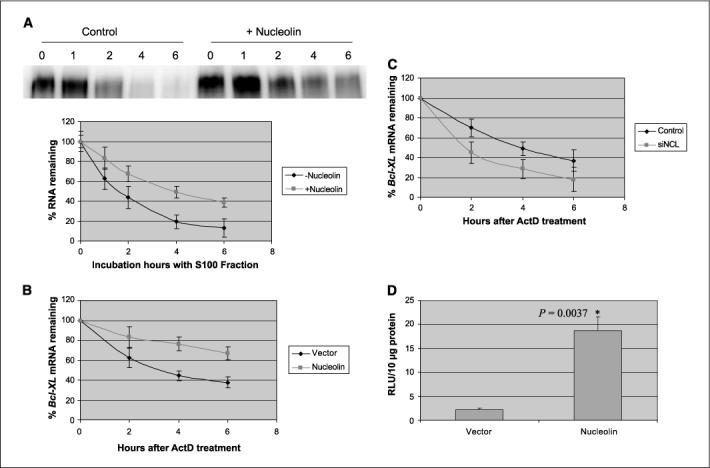
Nucleolin stabilizes the Bcl-XL mRNA in vitro and in vivo. A, 5 μg of the recombinant nucleolin was incubated with 32P-labeled Bcl-XL mRNA and S100 fraction of the cell. Samples were taken at the indicated time points for autoradiography. Top, a representative graph of in vitro mRNA decay assay; bottom, the quantitative illustration of the Bcl-XL degradation curve. Points, means of three independent experiments; bars, SD. B, HeLa cells were transfected with either empty vector or pEGFP-nucleolin. After 20 h, actinomycin D was added into the medium (5 μg/mL). The cells were harvested at 0, 2, 4, and 6 h after actinomycin D treatment for RNA preparation and reverse transcription. Real-time PCR was performed to quantitate the Bcl-XL mRNA. The Bcl-XL mRNA of each sample was normalized to the GAPDH mRNA. Points, means of three independent experiments; bars, SD. C, HeLa cells were transfected with nucleolin-specific siRNA or a control siRNA. After 60 h, actinomycin was added to the medium (5 μg/mL). The cells were harvested at 0, 2, 4, and 6 h after actinomycin D treatment for RNA preparation and reverse transcription. Real-time PCR was performed to quantitate the Bcl-XL mRNA. The Bcl-XL mRNA of each sample was normalized to the GAPDH mRNA. Points, means of three independent experiments; bars, SD. D, HeLa cells were cotransfected with the Bcl-XL 3′-UTR reporter and pEGFP-nucleolin or the empty vector. After 20 h, the cells were harvested for Renilla luciferase assay. Points, means of three independent experiments; bars, SD. The data were analyzed by two-sample t test with equal variations.
To examine whether nucleolin can also stabilize the mRNA in living cells, we compared the half-lives of the Bcl-XL mRNA in HeLa cells with gain-of-function or loss-of-function of nucleolin. HeLa cells have similar responses to UV irradiation and are easier to transfect. In comparison, the transfection efficiency is very low in HaCaT cells.
We transfected HeLa cells with pEGFP-nucleolin or the empty control vector pEGFPC1. There was ∼40% to 50% of the cells expressing green fluorescence 20 h after transfection (data not shown). We found that overexpression of Bcl-XL does extend the half-life of the Bcl-XL mRNA to longer than 6 h (Fig. 4B).
To support the above result, we also used siRNA to knockdown nucleolin expression. We found that there was a 60% reduction of nucleolin protein after 60 h of the siRNA treatment compared with the negative control (data not shown). Therefore, we compared the half-lives of the Bcl-XL mRNA in these cells. As shown in Fig. 4C, there was a reduction of the Bcl-XL mRNA in the nucleolin siRNA-treated cells.
In Fig. 3C, we showed that nucleolin binds to the 3′-UTR of the Bcl-XL mRNA. We reasoned that this binding is important for the nucleolin regulation of the Bcl-XL mRNA. To test this idea, we cotransfected the cells with nucleolin and luciferase reporter fused to the Bcl-XL 3′-UTR. We found that cotransfection with nucleolin increased the luciferase activity to ∼9-fold compared with the empty vector (pEGFPC1) control (Fig. 4D), indicating that the 3′-UTR may confer nucleolin stabilization of the Bcl-XL mRNA.
Poly(A) tail is required for the nucleolin stabilization of Bcl-XL
It has been an intriguing goal to understand how AREs and RNA-binding proteins modulate stability of their targets. There are two possibilities for nucleolin stabilization of Bcl-XL mRNA. One is that nucleolin binds to a certain region and protects that region from degradation. Alternatively, nucleolin may help protect the mRNA poly(A) tail from deadenylation, which is the first step for mRNA degradation. To distinguish the two possibilities, we devised a simple experiment. We synthesized the mRNA with or without the poly(A) tail and compared the nucleolin stabilization effects on these two RNA species. We reasoned that for the first possibility, nucleolin would have the same effect on the RNA stability using both mRNA species. However, the half-lives of the two mRNA species were different, indicating that nucleolin failed to stabilize the mRNA without polyadenylation (Fig. 5A). These results supported the second possibility that nucleolin might help protect the poly(A) tail from deadenylation.
Figure 5.
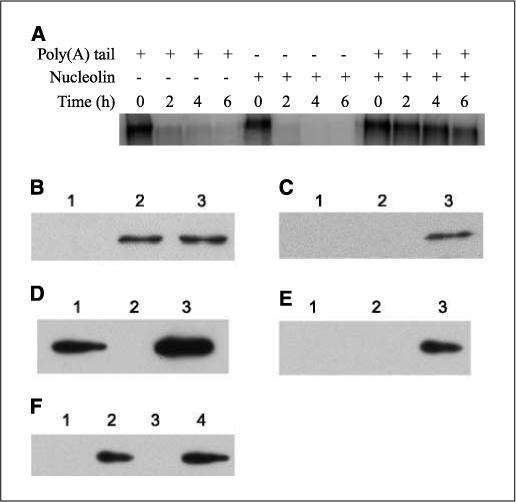
Nucleolin and PABP show physical interactions in vitro. A, nucleolin stabilization of the Bcl-XL mRNA is dependent upon the poly(A) tail. The 32P-labeled Bcl-XL mRNA with or without poly(A) tail was incubated with S100 fraction in the presence or absence of the recombinant nucleolin. The samples taken at indicated time points were subject to autoradiography. B-E, 1 μg of recombinant nucleolin (Nuc-RBD1,2,3,4-RGG9) and His6-PABP were incubated on ice for 1 h and then mixed with either amylase resin or Ni-NTA for 2 h at 4°C. The beads were washed, and the captured proteins were examined by Western blot analysis. B, Western blot analysis of nucleolin with the samples from amylase resin. C, Western blot analysis of nucleolin with the samples from Ni-NTA beads. D, Western blot analysis of PABP with the samples from Ni-NTA. E, Western blot analysis of PABP with the samples from amylase resin. Lane 1, PABP; lane 2, nucleolin; lane 3, PABP + nucleolin. F, 1 μg of the recombinant Nuc-RGG4 (lane 1) or Nuc-RGG9 (lane 2) or MBP (lane 3) was incubated with 1 μg of the recombinant PABP on ice for 1 h and then mixed with amylase resin for 2 h at 4°C. The beads were washed, and the proteins were analyzed with the anti-PABP antibody. Lane 4, 0.1 μg of PABP loaded on to gel directly as the positive control.
To rule out the possibility that the differential stabilization resulted from the poly(A) per se, the polyadenylated mRNA was incubated with S100 in the absence of nucleolin. As shown in Fig. 5A, this mRNA was degraded rapidly. Therefore, it was nucleolin, but not poly(A) tail, that stabilized RNA in this assay. In summary, this result also showed that the nucleolin stabilization effect was dependent upon poly(A) tail.
Nucleolin has a physical interaction with PABP
As the poly(A) tail is important for the nucleolin stabilization of Bcl-XL mRNA and the poly(A) tail is primarily bound by the PABP, we reasoned that this stabilization might be achieved through a physical interaction between nucleolin and PABP. To test this hypothesis, we used the purified recombinant proteins to examine their interactions. We reasoned that if they interacted with each other, pulling down either one of them could capture the other by affinity chromatography. Therefore, we used amylase resin to pull down MBP-tagged nucleolin and Ni-NTA agarose for His6-tagged PABP. As shown in Fig. 5B, nucleolin was detected in amylase resin (lanes 2 and 3) as expected. In Ni-NTA agarose, however, nucleolin was detected only when incubated with His6-PABP (Fig. 5C, lane 3). In comparison, nucleolin did not bind to the Ni-NTA beads (Fig. 5C, lane 2). Conversely, PABP did bind to Ni-NTA agarose (Fig. 5D, lanes 1 and 3) as expected, but it could only be detected in the amylose resin when incubated with MBP-nucleolin (Fig. 5E, lane 3). These results indicated that PABP and nucleolin interact with each other in vitro, and this interaction is not RNA dependent.
Nucleolin has four RNA-binding domains and nine Arg-Gly-Gly (RGG) motifs (43) at its COOH terminus. It has been shown that these RGG motifs are involved in interacting with both ribosomal proteins (55) and DNA G-quadraplex (43). Using the recombinant nucleolin with different domains, we found that the recombinant protein Nuc-RGG9 alone was able to pull down PABP (Fig. 5F, lane 2) even in the absence of any RNA-binding domains, whereas the truncated region Nuc-RGG4 failed to do so (Fig. 5F, lane 1). This result is similar to the previous report that the binding of Nuc-RGG9 to DNA G-quadraplex was abolished when Nuc-RGG9 was truncated to Nuc-RGG4 (43). Therefore, our data suggested that the full-length RGG motifs are required for interacting with PABP and the RNA binding domains may not be necessary for this interaction. Because the MBP fusion proteins were used in the experiments, we examined if MBP alone could interact with PABP. As shown in Fig. 5F (lane 3), there was no PABP detected. This result ruled out the possibility that MBP interacts with PABP.
To test if nucleolin interacts with PABP in intact cells, we used reciprocal immunoprecipitation to pellet both proteins. As expected, nucleolin was detected in the sample precipitated with the anti-PABP antibody (Fig. 6A), whereas the control IgG did not pellet nucleolin. Similarly, PABP was present in the antinucleolin antibody–precipitated sample (Fig. 6B). Therefore, these data strongly showed that PABP interacts with nucleolin.
Figure 6.
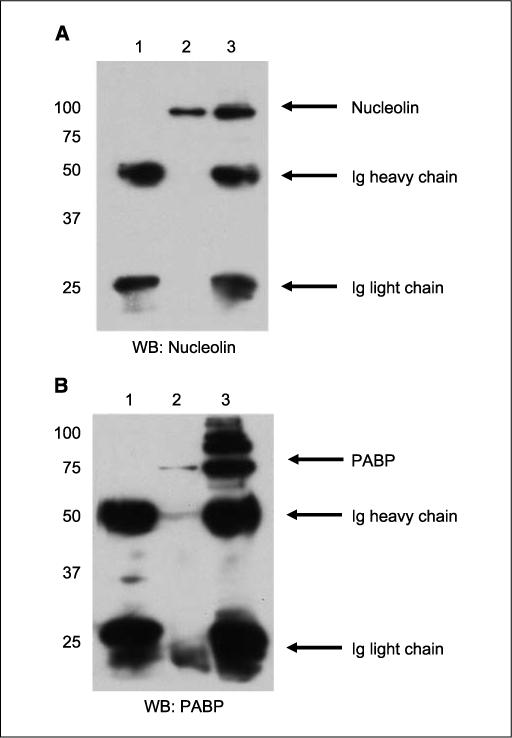
Nucleolin and PABP interact in vivo. Reciprocal immunoprecipitation of nucleolin and PABP were performed. A and B, lane 1, immunoprecipitation (IP) with control mouse IgG; lane 2, immunoprecipitation with antinucleolin antibody; lane 3, immunoprecipitation with anti-PABP antibody. A, immunoblotting with antinucleolin antibody; B, immunoblotting with anti-PABP antibody.
Discussion
The antiapoptotic molecule Bcl-XL has been implicated in tumor initiation, malignancy, and drug resistance (17, 20, 21, 56). Its importance in skin carcinogenesis has also been illustrated in both cell cultures and mouse models (18, 19, 22). Therefore, a better understanding of its regulation may shed light on novel strategies for skin cancer prevention and treatment. In this study, we provide evidence that nucleolin can stabilize the Bcl-XL mRNA by binding to the ARE elements of the 3′-UTR in vitro. Furthermore, we show that this stabilization effect is dependent upon the presence of poly(A) tail. This mechanism is similar to the stabilization of Bcl-2 mRNA, suggesting that there might be a conserved regulatory mechanism between these two mRNAs (37).
To better understand the molecular mechanism whereby the Bcl-XL mRNA is stabilized upon UVA irradiation, we used RNA affinity chromatography to capture proteins that were differentially bound to the Bcl-XL 3′-UTR. This technique has been successfully used in identifying RNA-binding proteins, i.e., Renin (57), and nucleoplasmin (51). In our experiment, we found that at least 10 protein bands were differentially bound to the RNA column upon UVA irradiation.
One of the bands was identified as nucleolin (Fig. 1). Nucleolin possesses four RNA-binding domains and reportedly stabilizes multiple RNA molecules (38, 41, 51). Interestingly, nucleolin stabilizes the Bcl-2 mRNA in HL-60 cells (37). Furthermore, we found that the binding of nucleolin to the beads increased in UVA-irradiated cells, indicating that nucleolin may play a role in the UVA response in human keratinocytes (Fig. 1, lane 2 and Fig. 3A, lane 3, top). Therefore, we reasoned that nucleolin might bind to and stabilize the Bcl-XL mRNA in UVA-irradiated human keratinocytes.
Nucleolin is a ubiquitously expressed protein and involved in important biological processes, such as cell cycle regulation and ribosomal biogenesis (58). Nucleolin has been implicated in apoptosis through various mechanisms. For example, overexpression of nucleolin increases the expression of p53 protein and thus promotes apoptosis in HCT116 or U2-OS cells by inhibiting MDM2-mediated ubiquitination (59). In contrast, nucleolin is antiapoptotic in other studies (37, 38, 51). Our data are consistent with previous findings that nucleolin regulates gene expression by stabilizing mRNAs (37, 38, 41). For example, nucleolin, together with another protein YB-1, stabilizes IL-2 mRNA in a JNK kinase–dependent manner (41). These two proteins recognize a cis-element called JNK response element at the 5′-UTR of IL-2 and stabilize IL-2 mRNA both in vitro and in vivo.
How nucleolin stabilizes the Bcl-XL mRNA remains unclear. However, in this report, we provide evidence that nucleolin stabilization of Bcl-XL requires the poly(A) tail and that there is a physical interaction between nucleolin and PABP. Our data suggest that nucleolin-PABP interaction may also be direct, because the recombinant proteins interact with each other in the absence of any RNA (Fig. 5). Furthermore, our data suggest that all RGG motifs of nucleolin may be required for this interaction (Fig. 5F).
The biological significance of the PABP-nucleolin interaction is not clear. A similar finding was reported. HuR, a well-studied RNA-stabilizing protein, was found to interact with PABP. Nagaoka et al. reported that HuR-PABP interaction is essential to the β-casein mRNA stability (40). Likewise, we postulate that interactions between PABP and RNA-stabilizing proteins, i.e., nucleolin, may block the action of PARN and therefore slow down deadenylation process. Furthermore, PABP has been shown to interact with proteins binding to the 5′-UTR, i.e., eIF, forming a RNA loop structure that stabilizes mRNA and facilitates translation. Therefore, we propose a similar model contributing to UVA-mediated resistance to apoptosis in human keratinocytes. In this model, UVA irradiation increases the binding capacity of nucleolin to the AREs on the 3′-UTR of Bcl-XL mRNA in a p38 MAPK-dependent manner. Subsequently, nucleolin binds to PABP in proximity through the RGG motifs, and this interaction helps stabilize the mRNA loop structure by excluding access of PARN and exosomes from digesting the poly(A) tail. Consequently, the Bcl-XL mRNA is stabilized and its expression increased.
This model does not exclude the possibility that nucleolin may also displace other destabilizing factors from the 3′-UTR. RNA-binding proteins may compete with same cis-elements. For example, HuR binding to the iNOS mRNA is mutually exclusive with KSRP binding (36). Similarly, HuR has been considered to compete with AUF1 (45). Finally, we have considered the possibility that p38-mediated phosphorylation of nucleolin prevents the recruitment of other destabilizing proteins, i.e., Tristetraprolin and KSRP, to mRNAs (35).
In this study, we provide evidence showing that nucleolin binds to the ARE of the 3′-UTR of the Bcl-XL mRNA and stabilizes the message both in vitro and in vivo. Because of the importance Bcl-XL in skin carcinogenesis, a better understanding of its regulation can be helpful for devising novel strategies for treatment and prevention of skin cancers.
Acknowledgments
Grant support: NIH grants CA23074, CA40584, and CA27502. Mass spectrometric data were acquired by the Arizona Proteomics Consortium supported by NIEHS grant ES06694 to SWEHSC, NIH/National Cancer Institute grant CA23074 to AZCC, and BIO5 Institute of the University of Arizona.
We thank Drs. Kiledjian, Maizels, and Kastan for providing reagents and Anne Cione for her administrative assistance.
References
- 1.Bode AM, Dong Z. Mitogen-activated protein kinase activation in UV-induced signal transduction. Sci STKE. 2003;2003:RE2. doi: 10.1126/stke.2003.167.re2. [DOI] [PubMed] [Google Scholar]
- 2.Bowden GT. Prevention of non-melanoma skin cancer by targeting ultraviolet-B-light signalling. Nat Rev Cancer. 2004;4:23–35. doi: 10.1038/nrc1253. [DOI] [PubMed] [Google Scholar]
- 3.Mitchell D. Revisiting the photochemistry of solar UVA in human skin. Proc Natl Acad Sci U S A. 2006;103:13567–8. doi: 10.1073/pnas.0605833103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Gruijl FR. Photocarcinogenesis: UVA vs UVB. Methods Enzymol. 2000;319:359–66. doi: 10.1016/s0076-6879(00)19035-4. [DOI] [PubMed] [Google Scholar]
- 5.de Laat JM, de Gruijl FR. The role of UVA in the aetiology of non-melanoma skin cancer. Cancer Surv. 1996;26:173–91. [PubMed] [Google Scholar]
- 6.He YY, Pi J, Huang JL, Diwan BA, Waalkes MP, Chignell CF. Chronic UVA irradiation of human HaCaT keratinocytes induces malignant transformation associated with acquired apoptotic resistance. Oncogene. 2006;25:3680–8. doi: 10.1038/sj.onc.1209384. [DOI] [PubMed] [Google Scholar]
- 7.Moan J, Dahlback A, Setlow RB. Epidemiological support for an hypothesis for melanoma induction indicating a role for UVA radiation. Photochem Photo-biol. 1999;70:243–7. [PubMed] [Google Scholar]
- 8.van Kranen HJ, de Laat A, van de Ven J, et al. Low incidence of p53 mutations in UVA (365-nm)-induced skin tumors in hairless mice. Cancer Res. 1997;57:1238–40. [PubMed] [Google Scholar]
- 9.de Laat A, van der Leun JC, de Gruijl FR. Carcinogenesis induced by UVA (365-nm) radiation: the dose-time dependence of tumor formation in hairless mice. Carcinogenesis. 1997;18:1013–20. doi: 10.1093/carcin/18.5.1013. [DOI] [PubMed] [Google Scholar]
- 10.Mouret S, Baudouin C, Charveron M, Favier A, Cadet J, Douki T. Cyclobutane pyrimidine dimers are predominant DNA lesions in whole human skin exposed to UVA radiation. Proc Natl Acad Sci U S A. 2006;103:13765–70. doi: 10.1073/pnas.0604213103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bachelor MA, Bowden GT. UVA-mediated activation of signaling pathways involved in skin tumor promotion and progression. Semin Cancer Biol. 2004;14:131–8. doi: 10.1016/j.semcancer.2003.09.017. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Y, Ma WY, Kaji A, Bode AM, Dong Z. Requirement of ATM in UVA-induced signaling and apoptosis. J Biol Chem. 2002;277:3124–31. doi: 10.1074/jbc.M110245200. [DOI] [PubMed] [Google Scholar]
- 13.Grad JM, Zeng XR, Boise LH. Regulation of Bcl-xL: a little bit of this and a little bit of STAT. Curr Opin Oncol. 2000;12:543–9. doi: 10.1097/00001622-200011000-00006. [DOI] [PubMed] [Google Scholar]
- 14.Kim R. Unknotting the roles of Bcl-2 and Bcl-xL in cell death. Biochem Biophys Res Commun. 2005;333:336–43. doi: 10.1016/j.bbrc.2005.04.161. [DOI] [PubMed] [Google Scholar]
- 15.Motoyama N, Wang F, Roth KA, et al. Massive cell death of immature hematopoietic cells and neurons in Bcl-x-deficient mice. Science. 1995;267:1506–10. doi: 10.1126/science.7878471. [DOI] [PubMed] [Google Scholar]
- 16.Wu LX, La Rose J, Chen L, et al. CD28 regulates the translation of Bcl-xL via the phosphatidylinositol 3-kinase/mammalian target of rapamycin pathway. J Immunol. 2005;174:180–94. doi: 10.4049/jimmunol.174.1.180. [DOI] [PubMed] [Google Scholar]
- 17.Kirkin V, Joos S, Zornig M. The role of Bcl-2 family members in tumorigenesis. Biochim Biophys Acta. 2004;1644:229–49. doi: 10.1016/j.bbamcr.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 18.Jost M, Class R, Kari C, Jensen PJ, Rodeck U. A central role of Bcl-X(L) in the regulation of keratinocyte survival by autocrine EGFR ligands. J Invest Dermatol. 1999;112:443–9. doi: 10.1046/j.1523-1747.1999.00543.x. [DOI] [PubMed] [Google Scholar]
- 19.Pena JC, Rudin CM, Thompson CB. A Bcl-xL transgene promotes malignant conversion of chemically initiated skin papillomas. Cancer Res. 1998;58:2111–6. [PubMed] [Google Scholar]
- 20.Chun E, Lee KY. Bcl-2 and Bcl-xL are important for the induction of paclitaxel resistance in human hepatocellular carcinoma cells. Biochem Biophys Res Commun. 2004;315:771–9. doi: 10.1016/j.bbrc.2004.01.118. [DOI] [PubMed] [Google Scholar]
- 21.Minn AJ, Rudin CM, Boise LH, Thompson CB. Expression of bcl-xL can confer a multidrug resistance phenotype. Blood. 1995;86:1903–10. [PubMed] [Google Scholar]
- 22.Schoop RA, Kooistra K, Baatenburg De Jong RJ, Noteborn MH. Bcl-xL inhibits p53- but not apoptin-induced apoptosis in head and neck squamous cell carcinoma cell line. Int J Cancer. 2004;109:38–42. doi: 10.1002/ijc.11675. [DOI] [PubMed] [Google Scholar]
- 23.Reed JC. Proapoptotic multidomain Bcl-2/Bax-family proteins: mechanisms, physiological roles, and therapeutic opportunities. Cell Death Differ. 2006;13:1378–86. doi: 10.1038/sj.cdd.4401975. [DOI] [PubMed] [Google Scholar]
- 24.Pecci A, Viegas LR, Baranao JL, Beato M. Promoter choice influences alternative splicing and determines the balance of isoforms expressed from the mouse bcl-X gene. J Biol Chem. 2001;276:21062–9. doi: 10.1074/jbc.M008665200. [DOI] [PubMed] [Google Scholar]
- 25.Viegas LR, Vicent GP, Baranao JL, Beato M, Pecci A. Steroid hormones induce bcl-X gene expression through direct activation of distal promoter P4. J Biol Chem. 2004;279:9831–9. doi: 10.1074/jbc.M312402200. [DOI] [PubMed] [Google Scholar]
- 26.Bachelor MA, Bowden GT. Ultraviolet A-induced modulation of Bcl-XL by p38 MAPK in human keratinocytes: posttranscriptional regulation through the 3′-untranslated region. J Biol Chem. 2004;279:42658–68. doi: 10.1074/jbc.M406626200. [DOI] [PubMed] [Google Scholar]
- 27.Grzybowska EA, Wilczynska A, Siedlecki JA. Regulatory functions of 3′UTRs. Biochem Biophys Res Commun. 2001;288:291–5. doi: 10.1006/bbrc.2001.5738. [DOI] [PubMed] [Google Scholar]
- 28.Shim J, Karin M. The control of mRNA stability in response to extracellular stimuli. Mol Cells. 2002;14:323–31. [PubMed] [Google Scholar]
- 29.Audic Y, Hartley RS. Post-transcriptional regulation in cancer. Biol Cell. 2004;96:479–98. doi: 10.1016/j.biolcel.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 30.Chen CY, Chen TM, Shyu AB. Interplay of two functionally and structurally distinct domains of the c-fos AU-rich element specifies its mRNA-destabilizing function. Mol Cell Biol. 1994;14:416–26. doi: 10.1128/mcb.14.1.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meyer S, Temme C, Wahle E. Messenger RNA turnover in eukaryotes: pathways and enzymes. Crit Rev Biochem Mol Biol. 2004;39:197–216. doi: 10.1080/10409230490513991. [DOI] [PubMed] [Google Scholar]
- 32.Mukherjee D, Gao M, O'Connor JP, et al. The mammalian exosome mediates the efficient degradation of mRNAs that contain AU-rich elements. EMBO J. 2002;21:165–74. doi: 10.1093/emboj/21.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stoecklin G, Mayo T, Anderson P. ARE-mRNA degradation requires the 5′-3′ decay pathway. EMBO Rep. 2006;7:72–7. doi: 10.1038/sj.embor.7400572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barreau C, Paillard L, Osborne HB. AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res. 2005;33:7138–50. doi: 10.1093/nar/gki1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chou CF, Mulky A, Maitra S, et al. Tethering KSRP, a decay-promoting AU-rich element-binding protein, to mRNAs elicits mRNA decay. Mol Cell Biol. 2006;26:3695–706. doi: 10.1128/MCB.26.10.3695-3706.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pan YX, Chen H, Kilberg MS. Interaction of RNA-binding proteins HuR and AUF1 with the human ATF3 mRNA 3′-untranslated region regulates its amino acid limitation-induced stabilization. J Biol Chem. 2005;280:34609–16. doi: 10.1074/jbc.M507802200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sengupta TK, Bandyopadhyay S, Fernandes DJ, Spicer EK. Identification of nucleolin as an AU-rich element binding protein involved in bcl-2 mRNA stabilization. J Biol Chem. 2004;279:10855–63. doi: 10.1074/jbc.M309111200. [DOI] [PubMed] [Google Scholar]
- 38.Zhang Y, Bhatia D, Xia H, Castranova V, Shi X, Chen F. Nucleolin links to arsenic-induced stabilization of GADD45α mRNA. Nucleic Acids Res. 2006;34:485–95. doi: 10.1093/nar/gkj459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rajagopalan LE, Westmark CJ, Jarzembowski JA, Malter JS. hnRNP C increases amyloid precursor protein (APP) production by stabilizing APP mRNA. Nucleic Acids Res. 1998;26:3418–23. doi: 10.1093/nar/26.14.3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nagaoka K, Suzuki T, Kawano T, Imakawa K, Sakai S. Stability of casein mRNA is ensured by structural interactions between the 3′-untranslated region and poly(A) tail via the HuR and poly(A)-binding protein complex. Biochim Biophys Acta. 2006;1759:132–40. doi: 10.1016/j.bbaexp.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 41.Chen CY, Gherzi R, Andersen JS, et al. Nucleolin and YB-1 are required for JNK-mediated interleukin-2 mRNA stabilization during T-cell activation. Genes Dev. 2000;14:1236–48. [PMC free article] [PubMed] [Google Scholar]
- 42.Hanakahi LA, Dempsey LA, Li MJ, Maizels N. Nucleolin is one component of the B cell-specific transcription factor and switch region binding protein, LR1. Proc Natl Acad Sci U S A. 1997;94:3605–10. doi: 10.1073/pnas.94.8.3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hanakahi LA, Sun H, Maizels N. High affinity interactions of nucleolin with G-G-paired rDNA. J Biol Chem. 1999;274:15908–12. doi: 10.1074/jbc.274.22.15908. [DOI] [PubMed] [Google Scholar]
- 44.Wang Z, Day N, Trifillis P, Kiledjian M. An mRNA stability complex functions with poly(A)-binding protein to stabilize mRNA in vitro. Mol Cell Biol. 1999;19:4552–60. doi: 10.1128/mcb.19.7.4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cok SJ, Acton SJ, Sexton AE, Morrison AR. Identification of RNA-binding proteins in RAW 264.7 cells that recognize a lipopolysaccharide-responsive element in the 3-untranslated region of the murine cyclooxygenase-2 mRNA. J Biol Chem. 2004;279:8196–205. doi: 10.1074/jbc.M308475200. [DOI] [PubMed] [Google Scholar]
- 46.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68:850–8. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 47.Andon NL, Hollingworth S, Koller A, Greenland AJ, Yates JR, III, Haynes PA. Proteomic characterization of wheat amyloplasts using identification of proteins by tandem mass spectrometry. Proteomics. 2002;2:1156–68. doi: 10.1002/1615-9861(200209)2:9<1156::AID-PROT1156>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 48.Yates JR, III, Eng JK, McCormack AL, Schieltz D. Method to correlate tandem mass spectra of modified peptides to amino acid sequences in the protein database. Anal Chem. 1995;67:1426–36. doi: 10.1021/ac00104a020. [DOI] [PubMed] [Google Scholar]
- 49.Cooper B, Eckert D, Andon NL, Yates JR, Haynes PA. Investigative proteomics: identification of an unknown plant virus from infected plants using mass spectrometry. J Am Soc Mass Spectrom. 2003;14:736–41. doi: 10.1016/S1044-0305(03)00125-9. [DOI] [PubMed] [Google Scholar]
- 50.Conne B, Stutz A, Vassalli JD. The 3′ untranslated region of messenger RNA: a molecular ‘hotspot’ for pathology? Nat Med. 2000;6:637–41. doi: 10.1038/76211. [DOI] [PubMed] [Google Scholar]
- 51.Yang C, Maiguel DA, Carrier F. Identification of nucleolin and nucleophosmin as genotoxic stress-responsive RNA-binding proteins. Nucleic Acids Res. 2002;30:2251–60. doi: 10.1093/nar/30.10.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Navaratnam N, Shah R, Patel D, Fay V, Scott J. Apolipoprotein B mRNA editing is associated with UV crosslinking of proteins to the editing site. Proc Natl Acad Sci U S A. 1993;90:222–6. doi: 10.1073/pnas.90.1.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hieronymus H, Silver PA. Genome-wide analysis of RNA-protein interactions illustrates specificity of the mRNA export machinery. Nat Genet. 2003;33:155–61. doi: 10.1038/ng1080. [DOI] [PubMed] [Google Scholar]
- 54.Hurt E, Luo MJ, Rother S, Reed R, Strasser K. Cotranscriptional recruitment of the serine-arginine-rich (SR)-like proteins Gbp2 and Hrb1 to nascent mRNA via the TREX complex. Proc Natl Acad Sci U S A. 2004;101:1858–62. doi: 10.1073/pnas.0308663100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bouvet P, Diaz JJ, Kindbeiter K, Madjar JJ, Amalric F. Nucleolin interacts with several ribosomal proteins through its RGG domain. J Biol Chem. 1998;273:19025–9. doi: 10.1074/jbc.273.30.19025. [DOI] [PubMed] [Google Scholar]
- 56.Krajewska M, Moss SF, Krajewski S, Song K, Holt PR, Reed JC. Elevated expression of Bcl-X and reduced Bak in primary colorectal adenocarcinomas. Cancer Res. 1996;56:2422–7. [PubMed] [Google Scholar]
- 57.Skalweit A, Doller A, Huth A, Kahne T, Persson PB, Thiele BJ. Posttranscriptional control of renin synthesis: identification of proteins interacting with renin mRNA 3′-untranslated region. Circ Res. 2003;92:419–27. doi: 10.1161/01.RES.0000059300.67152.4E. [DOI] [PubMed] [Google Scholar]
- 58.Srivastava M, Pollard HB. Molecular dissection of nucleolin's role in growth and cell proliferation: new insights. FASEB J. 1999;13:1911–22. [PubMed] [Google Scholar]
- 59.Saxena A, Rorie CJ, Dimitrova D, Daniely Y, Borowiec JA. Nucleolin inhibits Hdm2 by multiple pathways leading to p53 stabilization. Oncogene. 2006;25:7274–88. doi: 10.1038/sj.onc.1209714. [DOI] [PubMed] [Google Scholar]