Abstract
A highly sensitive electrochemical DNA hybridization assay using Prussian blue (PB)-modified polymeric spheres as the oligonculeotide labeling tag is described. The sandwich assay relies on a secondary nucleic-acid probe functionalized with polystyrene beads loaded with numerous Prussian blue nanoparticles. The very strong catalytic activity of the captured PB ‘artificial peroxidase’ tag towards the reduction of hydrogen peroxide, along with the encapsulation of numerous catalytic particles onto polymeric beads, allows amperometric detection of the DNA target down to the 50 fM level (2.5 amol). Imaging and spectroscopic measurements are used to characterize the PB-tagged polystyrene beads. Such coupling of PB catalytic labels with polymeric carrier beads offers great promise for amplified transduction of different biomolecular interactions.
Keywords: DNA hybridization, Prussian blue, Amplification, Microspheres
1. Introduction
The development of electrochemical DNA biosensors has been the subject of considerable effort [1 – 3]. Such devices offer great promise for nucleic acid assays with features that include high sensitivity, inherent miniaturization, low cost, independence of sample turbidity or optical path length, minimal power demands, and high compatibility with modern microfabrication technologies. Electrochemical hybridization biosensors commonly rely on the conversion of the base-pair recognition event into a useful current signal [1 – 3]. Such electronic transduction of DNA hybridization events has commonly been achieved in connection to electroactive indicators, enzyme tracers or nanoparticle tags [1 – 3].
Achieving very high sensitivity is a major goal in electrochemical detection of DNA hybridization. This commonly requires innovative approaches that couple different amplification platforms and amplification processes [4]. While the use of enzyme tags has been extremely attractive for this purpose, enzyme-based DNA assays often suffer from shortcomings associated with the limited stability and cost of the biocatalyst. Recent work by Polsky et al. [5] illustrated that nanoparticle tags can be used as catalytic labels, in a manner analogous to the use of enzyme tracers. For example, nucleic-acid functionalized platinum particles were shown to be useful for amplifying the detection of DNA hybridization down to the 10 pM level.
Here we report on an attractive method for amplifying the electrochemical detection of biomolecular interactions based on nucleic-acid polystyrene (PS) beads modified with Prussian-blue (PB) particles. PB is well recognized as the most attractive electrocatalyst for the reduction of hydrogen peroxide [6]. The favorable catalytic behavior of PB resembles that of commonly used peroxidase tags, and hence PB has often been denoted as ‘artificial enzyme peroxidase’ [7]. Such use of catalytic ‘artificial enzyme’ labels also addresses stability and cost limitations inherent to the use of enzyme tags.
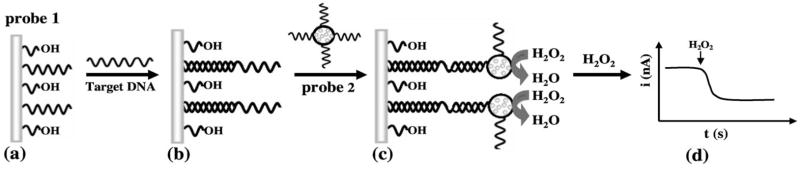
In the present study, we coupled the powerful catalytic action of PB with the amplification features of polymeric ‘carrier’ spheres [4, 8], by embedding PB nanoparticles within polystyrene (PS) spheres (linked to the secondary DNA probe). To our knowledge, the modification of polymeric spheres with catalytic PB nanoparticles has not been reported. The loading of PB particles onto gold nanoparticles was shown recently to offer high catalytic activity towards the reduction of hydrogen peroxide [9], but not in connection to bioaffinity assays. Figure 1 outlines the steps of the new microsphere-based bioelectronic sandwich assay. It involves co-immobilization of the DNA probe with 6-mercapto-1-hexanol on a gold surface (a), hybridization of the target DNA (b), binding of a secondary probe conjugated to PB-embedded PS spheres (c), and catalytic amperometric detection of hydrogen peroxide at the captured PS-PB spheres (d). As illustrated in the following sections, such coupling of polymeric ‘carrier’ beads with the strong catalytic action of the PB ‘artificial enzyme’ leads to a remarkably sensitive bioelectronic detection of DNA hybridization down to the amol level.
Fig. 1.
Schematic representation of the analytical protocol. a) Formation of a mixed monolayer of the thiolated DNA probe (probe 1) and 6-mercapto-1-hexanol on the gold electrode. b) Hybridization of probe 1 with the target DNA. c) Hybridization of the captured DNA target with the PS-PB beads-labeled probe 2. d) Amperometric detection of hydrogen peroxide at the captured PB-loaded particles.
2. Experimental
2.1. Apparatus
Electrochemical measurements were performed with an AutoLab Potentiostat (Eco Chemie, Netherlands), controlled by the GPES software. Scanning electron microscopy (SEM) images of the PS and PS-PB beads were obtained with an XL30 SEM instrument (FEI Co., Hillsboro, OR). Ultraviolet-visible (UV-vis) absorption spectrums were recorded in a 1 mm path-length cell coupled to a UV-2501 PC spectrophotometer (Shimadzu Scientific Instruments, Inc, CA). Fourier Transform Infrared (FT-IR) spectra were recorded using a Nicolet 6700 FT-IR (Thermo-Fisher, Madison, WI).
2.2. Reagents
Potassium ferrocyanide (K4Fe(CN)6), iron(III)chloride (FeCl3), potassium chloride (KCl), morpholineethanesulfonic acid (MES), N-(3-dimethylaminopropyl)-N-ethylcarbodiimide hydrochloride (EDC) and hydrogen peroxide were purchased from Sigma (St. Louis, MO). N-hydroxysulfosuccinimide (NHS) was obtained from Fluka (Buchs, Switzerland). Carboxylated polystyrene beads (1.04 μm diameter) were obtained from Bangs Laboratories (catalog number PC04N, Fishers, IN). DNA oligonucleotides were obtained from IDT Technologies (Corabille, IA). The following oligonucleotides were used:
| Probe 1: | 5′-SH-GAC CTA GTC CTT CCA ACA GC-3′ |
| Probe 2: | 5′-GGG TTT ATG AAA AAC ACT TTT TTT TT-NH2-3′ |
| Target: | 5′-AAA GTG TTT TTC ATA AAC CCA TTA TCC AGG ACT GTT TAT AGC TGT TGG AAG GAC TAG GTC-3′ |
| Noncomplementary DNA: | 5′-TTC CTT AGC CCC CCC AGT GTG CAA GGG CAG TGA AGA CTT GAT TGT ACA AAA TAC GTT TTG-3′ |
All chemicals were analytical reagent grade and all solutions were prepared with double-distilled deionized water.
2.3. Preparation of Prussian Blue Nanoparticles
Prussian blue nanoparticles (ca. 15 nm in diameter) were prepared based on the method of Miao et al. [9]. Briefly, seven-milliliter of deionized water, 1 mL of 10 mM KCl and 1 mL of 2 mM K4Fe(CN)6 were mixed with vigorous stirring. Then, 1 mL of 2 mM FeCl3 was slowly added to the mixture for 40 min. The resulting dark-blue colloidal solution was centrifuged for 20 min at 13,000 rpm and the nanoparticles were collected in the precipitate.
2.4. Encapsulation of the PB Nanoparticles within the PS Beads
The PB nanoparticles were encapsulated into the PS beads by mixing rapidly 5 mg of the beads in 400 μL of a chloroform/butanol solvent mixture (15%/85% v/v). Six mg of PB nanoparticles were then added to the mixture, allowing the encapsulation to proceed for 40 min. The beads were then washed with 50 mM phosphate buffer (pH 6.0).
2.5. Conjugation of the PB-PS Beads with the Probe 2 DNA
The PB-PS beads were incubated in 100 μL of 0.1 M NHS/0.4 M EDC (in 0.1 M MES buffer, pH 5.5) solution, for 30 min and centrifuged at 9000 rpm for 5 min. The beads were then incubated with 100 μL of a 4 μM 3′NH2 – DNA (probe 2) solution for 2 hours. Subsequently, the DNA-linked PS-PB beads were centrifuged at 3000 rpm for 3 min and washed with 50 mM phosphate buffer (pH 6.0) twice and then suspended in 300 μL of the same phosphate buffer.
2.6. Immobilization, Sandwich Hybridization and Detection
2.6.1. Immobilization of DNA Probe (Probe 1) on the Gold Electrode
The surface of the gold disk electrode (2 mm in diameter; CH Instruments, Austin, TX) was cleaned by exposure to Piranha solution for 20 min and then rinsed with water. (Safety note: the Piranha solution should be handled with extreme caution.) The electrode was polished sequentially using alumina slurries of 3, 1 and 0.5 μm (on a polishing pad). Its potential was then cycled between −0.3 to 1.5 V (vs. Ag/AgCl) in 1 M H2SO4 until a stable background voltammogram, typical of a clean gold surface, was observed. The oligonucleotide probe was immobilized onto the gold surface by exposing overnight the electrode to 50 μL of a 1 μM thiolated-oligonucleotide probe 1 solution in 0.05 M phosphate buffer (pH 7.4). The DNA modified gold surface was then treated for 20 min in 0.1 M solution of 6-mercapto-1-hexanol (dissolved in water/ethanol, 1/4 (v/v)) followed by rinsing with deionized water.
2.6.2. Sandwich DNA Hybridization
The oligonucleotide-probe modified gold electrode surface was incubated for 1 hour with 50 μL of the target DNA solution (in the hybridization buffer: 750 mM NaCl, 150 mM sodium citrate). The surface was then washed with the same buffer to remove the unbound DNA. Subsequently, the electrode was incubated for 1 hour with 50 μL of a solution containing the secondary DNA-linked PS-PB beads (0.83 mg/μL) and then washed with the washing buffer (50 mM Tris-HCl, 0.1% Tween 20, pH 7.4).
2.6.3. Electrochemical Detection
The three electrode system consisted of the DNA-modified gold working electrode, an Ag/AgCl reference electrode and a platinum wire counter electrode. The electrochemical response to the 10 mM hydrogen peroxide addition was monitored amperometrically at a fixed potential of 0.0 V (vs. Ag/AgCl) using an 3 mL cell containing a 50 mM phosphate buffer/0.1 M KCl (pH 6.0) solution.
2.7. Modification of the Gold Electrode with the PS-PB Layer
A 20 μL droplet of the PS-PB beads solution (5 mg of beads in 300 μL of 50 mM phosphate buffer pH 6.0) was placed on the gold disk electrode surface and allowed to dry. Then the surface of electrode was covered with 3 μL of a 3% Nafion solution.
3. Results and Discussion
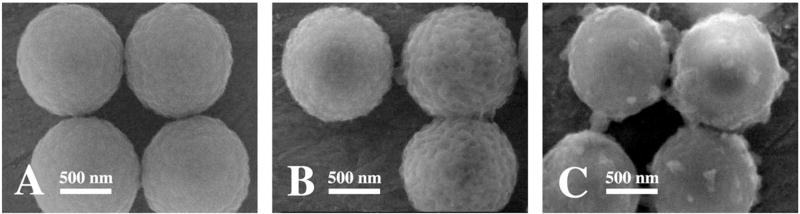
PB nanoparticles were prepared first in the reaction of the complex anion with the Fe3+ cation [9]. The PB particles were embedded into the PS microspheres during the swelling of the beads with organic solvent. The PB-embedded PS were characterized by scanning electron microscopy (SEM) and spectroscopic (UV-vis and FTIR) measurements. Figure 2 compares SEM images of the PS microspheres before (A) and after (B) swelling in the butanol/chloroform solution, as well as of the resulting PS-PB beads (C). While the initial surface of the PS beads is smooth, high degree of roughness is observed for the treated surface (A vs. B), reflecting the increased porosity associated with the swelling process. A similar treatment in the presence of the PB nanoparticles leads to a much smoother surface (C). This surface contains randomly distributed nonuniform globular microstructures corresponding to the Prussian blue. Such structures are not observed in the control experiment (without PB, B). Distinct PB nanoparticles cannot be observed under the low resolution of the SEM imaging.
Fig. 2.
SEM images of PS beads before (A) and after (B) swelling in a butanol/chloroform (85/15%) solution, as well as of the resulting PS-PB beads(C). SEM parameters include an accelerating voltage of 30 kV with 50 000× magnification.
The incorporation of PB into the PS spheres is indicated also from the UV-vis absorption spectra of Figure 3. The (naked) PS spheres display a broad absorption band around 400 nm (a). PB nanoparticles, in contrast, yield a single band around 710 nm (b), similar to that reported by Miao [9]. The spectra of the PS-PB beads (c) show two absorption signals, around 400 and 710 nm, characteristic of both the PS and PB, respectively. The FTIR spectra of the PS-PB spheres (not shown) displayed a new band around 2100 nm (compare to PS beads alone), characteristic of CN stretching [10]. The influence of the PB loading (in the ‘encapsulation’ solution) was examined from the amperometric response to hydrogen-peroxide of a PS-PB modified gold electrode (Fig. 4A). The peroxide current increases rapidly with the level of PB in the 400 μL solution between 2 and 6 mg and levels off thereafter. All subsequent work was carried out using a 6 mg PB loading solution. On-going studies examine the surface loading and coverage of PB on (and within) the PS spheres, and the extent of coverage over the surface vs. internal encapsulation within the swelling-induced nanopores.
Fig. 3.
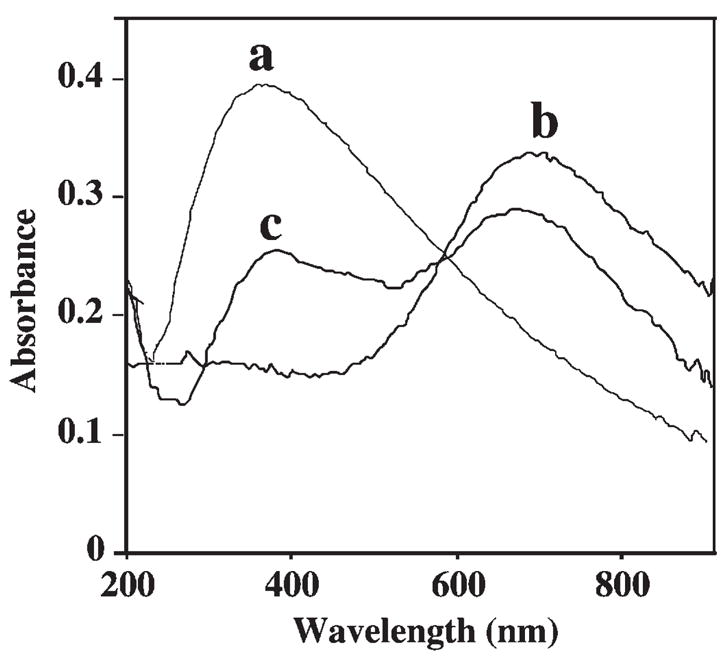
UV-vis absorption spectra of PS spheres (a), of PB nanoparticles (b), and of the PS-PB beads (c). Spectra were recorded in 50 mM phosphate buffer (pH 6.0).
Fig. 4.
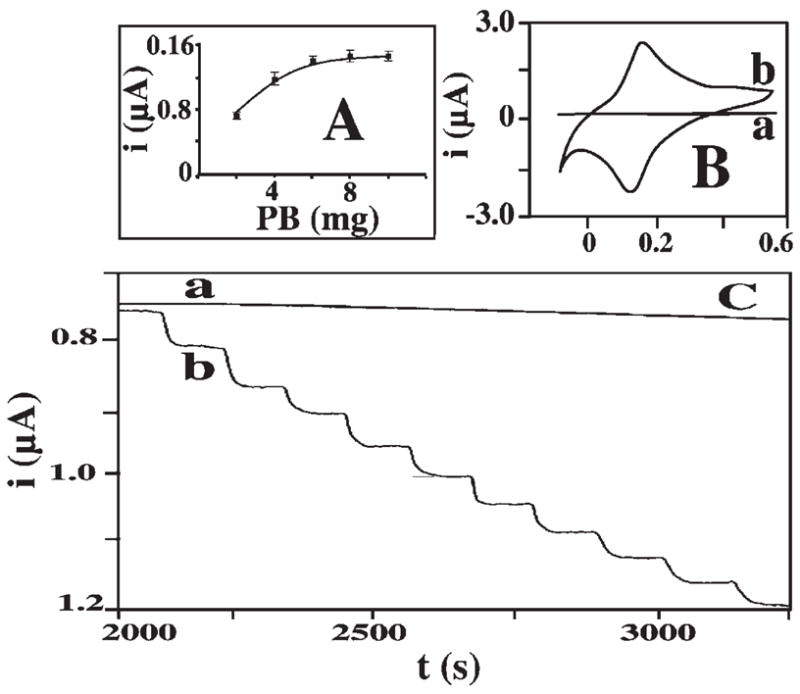
Electrocatalytic behavior of the PS-PB modified electrode. A) Effect of the loading of the PB particles on the PS beads on the amperometric response to 0.1 mM hydrogen peroxide. B) Cyclic voltammograms for 0.5 mM hydrogen peroxide at the PS-(a) and PS-PB (b) modified electrodes. C) Amperometric response to successive additions of 0.1 mM hydrogen peroxide at the unmodified (a) and PS-PB modified (b) gold electrodes. Operating potential (A), 0.0 V; scan rate (B), 50 mV/s. Electrolyte, 0.1 M KCl. Surface modification protocol. see Section 2.7.
The strong electrocatalytic activity of the PS-PB beads towards hydrogen peroxide was illustrated by using the catalytic spheres for modifying the surface of a gold-disk electrode (Fig. 4, B and C). The PS-PB modified electrode offers well-defined voltammetric (B) and amperometric (C) signals for hydrogen peroxide. Figure 2B compares cyclic voltammograms for hydrogen peroxide recorded at PS-(a) and PS-PB (b) sphere modified electrode. As expected, no peroxide response is observed over the entire −0.1 to +0.5 V potential range in the absence of the embedded PB particles. In contrast, a well-defined voltammogram, characteristic of PB modified electrodes [6], is observed using the PS-PB coating (ΔEp of 40 mV, Ep, = 130 mV, Ep, a = 170 mV). The resulting PS-PB modified electrode offers a sensitive low-potential amperometric detection of hydrogen peroxide. Figure 4C shows typical current-time recordings for 100 μM additions of hydrogen peroxide obtained at 0.0 V (vs. Ag/AgCl) using the PS-(a) and PS-PB (b) modified electrodes. The latter responds rapidly (ca. 20 s) to these peroxide additions and offers very favorable signal-to-noise characteristics. In contrast, no response is observed at the PS-modified surface (a), reflecting the absence of the PB catalyst. The PS-PB beads modified electrode also exhibited high operational stability, as was indicated from its highly stable response for hydrogen peroxide over 30 successive runs (not shown).
Subsequent DNA hybridization assays have relied on a secondary nucleic-acid probe functionalized with the PS-PB microspheres and electrocatalytic detection of hydrogen peroxide by the captured PS-PB particles. Such use of PS-PB catalytic labels for amplifying the detection of DNA hybridization is illustrated in Figure 5. This figure displays amperometric hybridization signals for extremely low target DNA concentrations ranging from 1 × 10−13 to 1 × 10−10 M (Fig. 5A, c – f). Well defined current signals are observed for these picomolar concentrations of the target. A detection limit of 5 × 10−14 M target DNA can be estimated based on the signal-to-noise characteristics (S/N =3) of the response for the 1 × 10−13 M target (c). Such detection limit corresponds to 46 fg (2.5 amol) in the 50 μL samples. The present protocol is thus ca. 100 times more sensitive compared to the DNA electronic detection based on platinum-nanoparticle catalytic labels [5]. A negligible signal is also observed in Figure 5 for a substantially (1000-fold) higher level of a noncomplementary oligomer (b). This small response is similar to that observed without the DNA target (a) and reflects primarily the negligible nonspecific adsorption of the probe-2/PS-PB conjugate. Such effective discrimination is attributed to hydrophilic character of the mixed monolayer of thiolated probe 1 and 6-mercapto-1-hexanol [11]. The corresponding calibration plot of current vs. log [DNA] (Fig. 5C) is linear over the 10−10 to 10−13 M range, with a correlation coefficient of 0.997. The amplified electrical signal is coupled to a relatively good reproducibility. The precision was estimated from a series of six successive trace measurements of 1 × 10−12 M target DNA that yielded reproducible current signals, with a RSD of 11%.
Fig. 5.
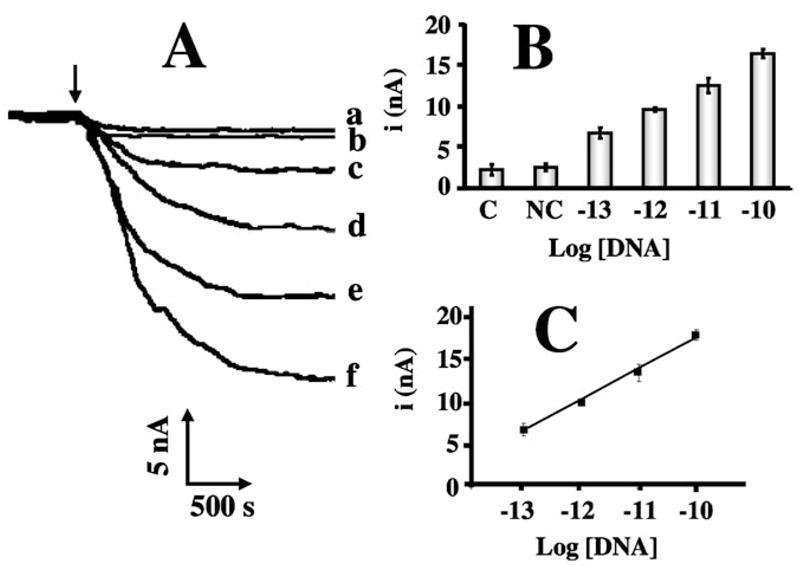
A) Amperometric response to different concentrations of the target DNA: 0 (a), 1 × 10−13 M (c), 1 × 10−12 M (d), 1 × 10−11 M (e) and 1 × 10−10 M (f), along with the response to 1 × 10−7 M of a noncomplementary DNA (b). B) Relative current signals for the different levels of the target DNA (shown in A). C) The corresponding semi-log calibration plot over the 1 × 10−10 – 1 × 10−13 M range of the DNA target. Hybridization buffer: 750 mM NaCl, 150 mM sodium citrate; hybridization time, 1 h. Electrochemical measurements were performed using a 0.1 M phosphate buffer/0.1 M KCl (pH 6.0) solution, a potential of 0.0 V, and additions of 10 mM hydrogen peroxide.
4. Conclusions
We have demonstrated for the first time the use of PS-PB spheres as tags for highly sensitive bioelectronic detection of DNA hybridization. Such tags couple the amplification features of polymeric carrier beads with the powerful catalytic action of the PB ‘artificial enzyme’. While the concept of PS-PB tags has been demonstrated for amplifying the transduction of DNA hybridization, it could be readily expanded for the monitoring of other target analytes such as proteins or glycans. The coupling of polymeric spheres with PB particles also holds great promise for surface coatings for improved oxidase-based enzyme electrodes and for enhancing the detection of different peroxide species ranging from hydrogen peroxide to peroxide explosives. Other catalytic labels (e.g., Pt nanoparticles [5]) could be coupled with the amplification features of polymeric carrier spheres and used for ultrasensitive monitoring of different biomolecular interactions.
Acknowledgments
This work was supported by the National Science Foundation (Grants CHE 0506529) and National Institutes of Health (Award Numbers 1U01AI075565, EB002189 and R01A 1056047-04). S. S. acknowledges a fellowship from the Thailand Research Fund (Royal Golden Jubilee Ph.D. Program).
Footnotes
Dedicated to Professor Ernö Pretsch on the Occasion of His Retirement from ETH Zürich
References
- 1.Palecek E, Fojta M. Anal Chem. 2001;73:75A. [Google Scholar]
- 2.Wang J. Anal Chim Acta. 2002;469:63. [Google Scholar]
- 3.Merkoci A. Electroanalysis. 2007;19:739. [Google Scholar]
- 4.Wang J. Small. 2005;1:1036. [Google Scholar]
- 5.Polsky R, Gill R, Kaganorsky L, Willner I. Anal Chem. 2006;78:2268. doi: 10.1021/ac0519864. [DOI] [PubMed] [Google Scholar]
- 6.Karyakin AA. Electroanalysis. 2001;13:813. [Google Scholar]
- 7.Karyakin AA, Karyakina EE, Gorton L. Anal Chem. 2000;72:1720. doi: 10.1021/ac990801o. [DOI] [PubMed] [Google Scholar]
- 8.Wang J, Polsky R, Merkoci A, Turner KL. Langmuir. 2003;19:989. [Google Scholar]
- 9.Miao Y, Chem J, Wu X, Miao J. Colloids Surf A, Physicochem Eng. 2007;295:135. [Google Scholar]
- 10.Kulesza PJ, Malik MA, Denca A, Strojek J. Anal Chem. 1996;68:2442. [Google Scholar]
- 11.Levicky R, Herne T, Tarlov M, Satija S. J Am Chem Soc. 1998;120:9787. [Google Scholar]