Abstract
We present here a genome-wide map of abnormalities found in diagnostic samples from 45 adults and adolescents with acute lymphoblastic leukemia (ALL). A 500K SNP array analysis uncovered frequent genetic abnormalities, with cryptic deletions constituting half of the detected changes, implying that microdeletions are a characteristic feature of this malignancy. Importantly, the pattern of deletions resembled that recently reported in pediatric ALL, suggesting that adult, adolescent, and childhood cases may be more similar on the genetic level than previously thought. Thus, 70% of the cases displayed deletion of one or more of the CDKN2A, PAX5, IKZF1, ETV6, RB1, and EBF1 genes. Furthermore, several genes not previously implicated in the pathogenesis of ALL were identified as possible recurrent targets of deletion. In total, the SNP array analysis identified 367 genetic abnormalities not corresponding to known copy number polymorphisms, with all but two cases (96%) displaying at least one cryptic change. The resolution level of this SNP array study is the highest used to date to investigate a malignant hematologic disorder. Our findings provide insights into the leukemogenic process and may be clinically important in adult and adolescent ALL. Most importantly, we report that microdeletions of key genes appear to be a common, characteristic feature of ALL that is shared among different clinical, morphological, and cytogenetic subgroups.
Keywords: deletions, genetic abnormality, t(9;22)
Acute lymphoblastic leukemia (ALL) occurs at all ages but displays a bimodal distribution of incidence, with one peak in early childhood and a second in patients older than 50 years (1). For adult ALL, the yearly incidence is ≈2 per 100,000, with ≈75% of the cases being B lineage and the remainder of T cell origin (1, 2). The most prominent prognostic factors are age, white blood cell count (WCC), and different genetic abnormalities, with younger age and lower WCC being associated with an improved outcome (3, 4). Most adults, however, are considered to be high-risk, and the long-term disease-free survival rates are <40% (1, 3–5). This is in stark contrast to pediatric ALL, where refined treatment regimens have resulted in cure rates approaching 80% (6, 7). Treatment of adolescents (15–21 years old) on pediatric protocols has resulted in an increased overall survival but is still far from the outstanding results achieved in children (8). Hence, there is a need for novel prognostic markers in adult and adolescent ALL to allow a better risk stratification of these patients and to identify new treatment targets.
Whereas genetic abnormalities in pediatric ALL are widely used in clinical practice for risk stratification purposes, most treatment protocols in adult ALL consider only the presence or absence of the Philadelphia chromosome, t(9;22)(q34;q11.2). The t(9;22), which results in the BCR/ABL1 fusion gene, is found in 20–30% of adult B cell precursor cases and is an adverse prognostic indicator (9–12). Other recurrent rearrangements include the t(4;11)(q21;q23) forming a MLL/AFF1 (previously AF4, MLLT2) fusion, the t(1;19)(q23;p13), which results in a fusion of TCF3 and PBX1, and the t(8;14)(q24;q32), which juxtaposes MYC and the Ig heavy chain locus, leading to overexpression of the former gene (13). However, the latter abnormalities are relatively rare (2–5% each), and their clinical impacts are still unclear, although most studies have reported an inferior outcome for t(4;11)- and t(8;14)-positive cases (9–12).
Although karyotype analysis has played an important role in the understanding of the pathogenesis of ALL, it is likely that submicroscopic, cytogenetically cryptic events are also involved in leukemogenesis. During recent years, the development of array-based comparative genome hybridization and SNP genotyping has enabled genome-wide detection of copy-number changes with a much higher resolution than can be acquired with standard cytogenetics (14). Several studies using these techniques have shown that submicroscopic imbalanced changes, which lead to a net gain or loss of genetic material, are common in hematologic malignancies, including acute myeloid leukemia (AML), myelodysplastic syndromes, and pediatric ALL (15–19). However, no such investigation has, as yet, focused on adolescent and adult ALL.
In the present study, we have used three different arrays for SNP genotyping, together giving a total of >500,000 SNPs with a median intermarker distance of <2.5 kb. We investigated a cohort of 45 adult and adolescent ALL cases, with the aim of identifying submicroscopic genetic anomalies. The resolution of the 500K system is one of the highest used to date to investigate a neoplastic disorder, and the present study uses SNP arrays to specifically address adult ALL. We here report that cryptic genetic changes are present in close to 100% of adult and adolescent ALL cases and show that, in line with recent findings in pediatric ALL (16, 17), intrachromosomal deletions of genes involved in B lymphopoiesis and cell-cycle regulation occur with a high frequency in this disorder. Furthermore, we identify gene targets that have not previously been implicated in ALL.
Results
Genome-Wide Screening of Leukemia-Associated Changes.
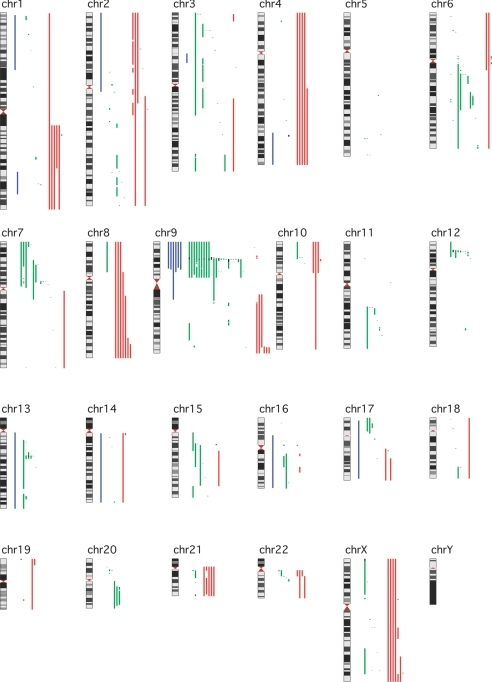
SNP array analysis, using a combination of three different arrays together comprising >500,000 SNPs, detected a total of 367 possible leukemia-related genetic changes among the 45 cases. These comprised 211 hemizygous deletions, 48 homozygous deletions, 93 copy-number gains, and 15 regions displaying uniparental disomy (UPD) [Fig. 1 and supporting information (SI) Dataset S1]. In addition, 57 previously described copy number polymorphisms (CNPs) [according to the Database of Genomic Variants, http://projects.tcag.ca/variation/ (20)] and 109 deletions because of somatic rearrangements in T cell receptor or Ig genes were identified; these were not analyzed further (data not shown). The median size of the 211 hemizygous deletions was 1.25 Mb (range 296 bp–129 Mb). The majority of the hemizygous deletions, 140 changes, was ≤5 Mb and hence expected to be cytogenetically cryptic. There were no monosomies. The median size of the homozygous deletions was 76.5 kb (range 189 bp–3.76 Mb); all would be expected to be cytogenetically cryptic. The median size of the 94 copy-number gains was 29.0 Mb (range 84.0 kb–246 Mb). Twenty gains involved whole chromosomes, whereas 25 were ≤5 Mb.
Fig. 1.
Overview of all genetic aberrations found with SNP array in 45 adult and adolescent ALL cases. Minimally involved regions are shown to the right of each chromosome. For each type of aberration, each line represents a different case. Blue lines are regions of uniparental disomy, light green lines are hemizygous deletions, dark green lines are homozygous deletions, and red lines are copy-number gains. Note the high frequency of deletions involving chromosomes 9p21.3, 9p13.2, 7p12.2, 12p13.2, and 13q14.2 corresponding to the CDKN2A, PAX5, IKZF1, ETV6, and RB1 loci, respectively.
The acquired UPDs comprised three whole-chromosome UPDs and 12 partial UPDs (pUPDs) (Dataset S1). The only region that recurrently displayed UPD was 9p, from ptel to 9p21.1. In five of six cases with such abnormalities, the partial UPD was associated with homozygous deletion of CDKN2A. In addition, five regions displaying >50 consecutive homozygous SNPs were detected in the same case (no. 15) and excluded from further analysis because they most likely were constitutional and due to consanguinity (data not shown).
The median number of aberrations per adult ALL case was six (range 0–40 aberrations); only two cases did not display any copy number change or UPD with SNP array analysis (Table 1).
Table 1.
Specific genes are targeted by deletions in adult ALL
| Case no. | Group | Total no of changes | LEF1 | EBF1 | IKZF1 | CDKN2A | PAX5 | DLG2 | ETV6 | RB1 | CDH13 | LDOC1 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Ph+ | 7 | HeD | HeD | ||||||||
| 2 | Ph+ | 13 | HoD | HoD | HeD | |||||||
| 3 | Ph+ | 20 | HeD | HoD | HeD | |||||||
| 4 | Ph+ | 13 | HeD | HoD | HeD | |||||||
| 5 | Ph+ | 19 | HeD | HoD | HeD | |||||||
| 6 | Ph+ | 11 | ||||||||||
| 7 | Ph+ | 4 | HeD | HeD | ||||||||
| 8 | Ph+ | 12 | HoD | HoD | HeD | |||||||
| 9 | Ph+ | 5 | HeD | |||||||||
| 10 | Ph+ | 12 | HoD | HeD | ||||||||
| 11 | 11q23 | 12 | ||||||||||
| 12 | 11q23 | 0 | ||||||||||
| 13 | t(8;14) | 4 | ||||||||||
| 14 | t(8;14) | 2 | ||||||||||
| 15 | Other | 7 | HeD | HoD | HeD | |||||||
| 16 | Other | 4 | HoD | HeD | ||||||||
| 17 | Other | 40 | HeD | HeD | HeD | HeD | ||||||
| 18 | Other | 17 | HeD | HoD | ||||||||
| 19 | NK | 7 | HoD | UPD | HoD | UPD | ||||||
| 20 | NK | 5 | HoD | |||||||||
| 21 | NK | 3 | ||||||||||
| 22 | NK | 4 | HeD | |||||||||
| 23 | N/A | 6 | HoD | HeD | ||||||||
| 24 | N/A | 3 | HeD | HeD | ||||||||
| 25 | N/A | 6 | HeD | |||||||||
| 26 | iAMP21 | 21 | HeD | HoD | ||||||||
| 27 | T-ALL | 4 | HoD | |||||||||
| 28 | T-ALL | 5 | UPD | UPD | ||||||||
| 29 | T-ALL | 2 | ||||||||||
| 30 | T-ALL | 1 | ||||||||||
| 31 | T-ALL | 2 | ||||||||||
| 32 | T-ALL | 5 | HoD | |||||||||
| 33 | T-ALL | 6 | HeD | HoD | ||||||||
| 34 | T-ALL | 6 | HeD | |||||||||
| 35 | T-ALL | 5 | HeD | |||||||||
| 36 | T-ALL | 5 | HoD | HoD | ||||||||
| 37 | T-ALL | 12 | HoD | HeD | HeD | |||||||
| 38 | T-ALL | 5 | HoD | HeD | ||||||||
| 39 | T-ALL | 7 | HeD | HoD | ||||||||
| 40 | T-ALL | 10 | HoD | |||||||||
| 41 | T-ALL | 3 | HeD | HeD | ||||||||
| 42 | N/A | 11 | HoD | HeD | HoD | |||||||
| 43 | N/A | 0 | ||||||||||
| 44 | N/A | 9 | HeD | |||||||||
| 45 | N/A | 12 | HeD | HeD |
Homozygous deletions (HoD), hemizygous deletions (HeD), or regions of uniparental disomy (UPD) for one or more of the LEF1, EBF1, IKZF1, CDKN2A, PAX5, DLG2, ETV6, RB1, CDH13, and LDOC1 genes were found in 35 of 45 cases (78%). The total number of leukemia-associated changes found in each case varied between 0 and 40. Ph+, t(9;22)-positive; NK, normal karyotype; iAMP21, intrachromosomal amplification of chromosome 21; and N/A, not available.
Recurrent Deletions Point to Novel Target Genes.
The analysis revealed 103 recurrent copy-number changes encompassing 75 deleted regions and 28 gained regions (Table S1). Several recurrently (in more than one case) lost or gained regions included genes previously implicated in leukemia such as MLL and MLLT10, although these could not be identified as definite targets because several other loci were in the deleted region. However, focal or overlapping deletions suggested previously uncharacterized single-gene targets for some chromosomal regions, e.g., DLG2 in four cases (8.9%), LDOC1 in three cases (6.7%), and CDH13 in two cases (4.4%) (Table 1, Fig. S1, and Table S1).
Deletions of Genes Involved in Cell-Cycle Regulation and B Lymphopoiesis.
Deletions of known leukemia-associated genes occurred with a high frequency. These included CDKN2A (P16) in 21 cases (47%), PAX5 in 15 cases (33%), IKZF1 (IKAROS) in eight cases (18%), ETV6 in seven cases (16%), RB1 in five cases (11%), EBF1 in two cases (4.4%), and LEF1 in one case (2.2%) (Table 1 and Table S1). Thirty-two of the 45 (71%) adult ALLs harbored a deletion of at least one of these genes. CDKN2A deletions were frequently homozygous (17 of 21 cases; 81%) and focal (<1 Mb in size; 13 of 21 cases; 62%). The homozygous CDKN2A deletions were either flanked by larger hemizygously deleted regions (12 of 17 cases; 71%) or were in regions of UPD (5 of 17 cases; 29%) (Fig. 2 and Table S2). PAX5 deletions were all hemizygous and frequently large; in all but three cases, the deleted region also included CDKN2A. The remaining genes displayed a mixture of hemi- and homozygous events.
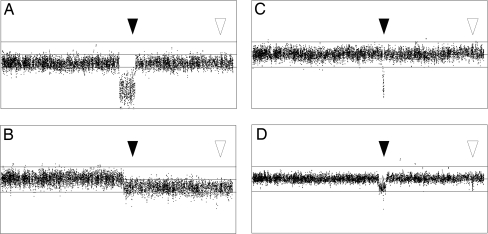
Fig. 2.
SNP array analysis results from the 250K Nsp and the 250K Sty arrays showing the 9p arm (1–39,401,290 bp) in four different cases. Each dot represents the log2 ratio of one SNP, with a moving average of three SNPs. The top line represents a log2 ratio of 1, the middle line a log2 ratio of 0, and the lower line a log2 ratio of −1. The positions of CDKN2A (black arrows) and PAX5 (white arrows) are shown. (A) Case 37, which has a hemizygous deletion of the entire 9p chromosome arm, including PAX5, and a 2.6-Mb homozygous deletion encompassing CDKN2A. (B) Case 41, which has a large hemizygous deletion starting at chr9; 20.6 Mb and encompassing CDKN2A and PAX5. (C) Case 39, which has a 208-kb homozygous deletion targeting CKDN2A associated with a partial uniparental disomy for the 9p arm. (D) Case 24, which has one 1.2-Mb hemizygous deletion and one 71-kb hemizygous deletion targeting CDKN2A and PAX5, respectively.
Clinical Correlations.
Deletion of RB1 correlated with older age (P = 0.011), and ETV6 deletions displayed a trend toward being significantly associated with younger age (P = 0.060). There was also a trend toward significance for IKZF1 deletion being associated with t(9;22) (P = 0.069) in B lineage ALL. There were no statistically significant gender, B/T cell lineage, age, or WCC-related differences for CDKN2A, PAX5, and IKZF1 and no gender, B/T cell lineage, or WCC-related differences for RB1 and ETV6.
Cytogenetic Abnormalities Detected by SNP Array Analysis.
Overall, the concordance between G banding/FISH and SNP array data were high regarding copy number for cases for which G banding karyotypes were available [35 of 45 (78%); Table S3]. Two cases could be reassigned to well known genetic subgroups based on SNP array results, despite normal G banding karyotypes. These were case 13, which displayed gain of 8q and loss of 14q with breakpoints adjacent to MYC and IGH suggesting a der(14)t(8;14)(q24.1;q32), and case 28, which harbored a 0.4-Mb deletion on chromosome 11, deleting the 3′ part of MLL and possibly resulting in a MLL fusion gene. In addition, case 26, for which no G banding karyotype was available, displayed a complex pattern of gains and losses in chromosome 21 and most likely belonged to the intrachromosomal amplification of chromosome 21 (iAMP21) karyotypic subgroup (21).
Gain of 1q was detected with SNP array analysis in 7 of 45 (16%) of the cases (Table S4). The minimally gained region was 1.36 Mb (chr1: 152.959805–154.321968 Mb) at 1q22. A total of 18 deletions involving 6q were found in 10 cases; seven different regions were deleted in three or more cases (Table S5).
There were a total of 14 balanced rearrangements [nine t(9;22), two t(4;11), one t(8;14), one inv(14), and one t(9;11)] found by G banding among the 45 adult ALLs (Table S3). Two of these cases, both t(9;22)-positive, harbored hemizygous microdeletions adjacent to the translocation breakpoints. Case 2 had a 2.0-Mb deletion on chromosome 9 (chr9: 130.649795–132.698182 Mb) including the 5′ part of ABL1 and a 324-kb deletion on chromosome 22 (chr22: 21.965924–22.290187 Mb) including the 3′ part of BCR, whereas case 6 had a 121-kb deletion on chromosome 9 (chr9: 132.594866–132.715588 Mb) in the 5′ part of ABL1 (Dataset S1).
Discussion
Genetic abnormalities are the driving force behind leukemogenesis and may be used for prognostication purposes as well as for identifying possible treatment targets, both much needed in adult ALL. In the present study, we have investigated 45 cases of adult and adolescent ALL using genome-wide SNP arrays comprising >500,000 SNPs, enabling the detection of submicroscopic copy-number changes and copy-neutral abnormalities.
We found a total of 367 putatively leukemia-associated genetic aberrations, with a median number of six abnormalities per case. Losses were more common than gains, and the deletions were frequently very small; microdeletions comprised more than half (53%) of the copy-number aberrations found in this study. Several possible target genes were identified, including DLG2 at 11q14.1 (four cases; 8.9%), LDOC1 at Xq27.2 (three cases; 6.7%), and CDH13 at 16q23.3 (two cases; 4.3%) (Table 1 and Fig. S1), based on focal deletions of only one locus or small minimally overlapping regions. Whereas DLG2 and LDOC1 have not been previously implicated in hematologic malignancies, methylation of the CDH13 promoter has been reported in 35% of pediatric and adult ALLs (22), suggesting that this gene may be silenced either by deletion or by methylation in ALL. However, the lack of constitutional DNA from the patients in this study means that we cannot definitively exclude that some of the detected changes may be CNPs, necessitating future studies to show whether DLG2, LDOC1, and CDH13 deletions are leukemogenic in adult ALL.
Another prominent finding in the present study was the frequent deletions of genes involved in cell-cycle regulation and in B lymphopoiesis (Table 1). These included CDKN2A (47% of the cases), PAX5 (33%), IKZF1 (18%), ETV6 (16%), RB1 (11%), EBF1 (4.4%), and LEF1 (2.2%). Deletions in 9p21.3 targeting CDKN2A are well known in adult ALLs (9, 23), although the high occurrence of homozygous deletions (17 of 21 cases with 9p21 loss; 81%) has not been previously reported. As regards ETV6, a recent FISH study reported ETV6 deletions in 3 of 74 (4%) of adult patients (24), which is substantially lower than the 18% detected among our cases, suggesting that SNP array analysis is in some instances superior to FISH for detection of small deletions. Deletions of PAX5, IKZF1, RB1, EBF1, or LEF1 have previously only been reported in single cases of adult ALL, but studies of pediatric cases have shown frequent loss (16, 17).
In fact, our findings in adult ALL are strikingly similar to what has recently been reported in pediatric ALL. Mullighan et al. (16) detected deletions of CDKN2A, PAX5, IKZF1, ETV6, RB1, EBF1, and LEF1 in a high proportion of childhood cases; a finding that was subsequently confirmed by Kuiper et al. (17). Kawamata et al. (25) also reported frequent CDKN2A and ETV6 deletions in pediatric cases, although they did not detect deletions of the other genes, either because of different sampling populations or differing analytical methods. Compared with the dataset from Mullighan et al. (16), which provide immunophenotype data, we found very similar frequencies for deletions of CDKN2A, PAX5, ETV6, and RB1 within the B and T immunophenotypic groups, although the fact that we included only 15 T-ALL cases in our study may preclude detection of small differences. For PAX5, which encodes a master regulator of B lymphopoiesis (26), all three cases with focal deletions were pre-B ALL, whereas both T-ALLs with loss of PAX5 harbored large deletions, including most of 9p, which is also in line with previous findings in pediatric ALL (16). The only gene that differed between adult and childhood cases was IKZF1, which was deleted in 27% (7 of 26) of the adult ALL cases of B cell lineage (Table 1), in comparison with 9% of pediatric ALL (P < 0.01) (16). Furthermore, when all our cases were analyzed together, RB1 deletions were associated with older age (P = 0.011), and all seven ETV6 deletions were found in patients 35 years of age or younger, suggesting that the frequency of these deletions may vary with age in adolescent and adult ALL.
The similarity between childhood and adult ALL as regards deletions was unexpected considering that the cytogenetic patterns of these disease entities are quite different—in particular, adult ALLs display a higher frequency of t(9;22)-positive cases but virtually lack the most common pediatric abnormalities, t(12;21) and high hyperdiploidy (9–12). Furthermore, several of the deletions were found in both B and T cell lineage ALL, including focal deletions of CDKN2A, ETV6, and RB1, suggesting that they may not be exclusive for one particular subgroup. This is in stark contrast to fusion gene-forming abnormalities, which are typically strongly associated with specific clinical and/or morphological features (27). It is thus possible that microdeletions in comparison may play a more general leukemia-promoting role and may be shared among different clinical, morphological and cytogenetic subgroups of ALL.
The SNP array analysis also detected well known leukemia-associated abnormalities, including gains of 1q in seven cases and seven recurrently deleted regions at 6q (Tables S4 and S5). Interestingly, the 1.4-Mb minimally gained 1q region included the gene DAP3, which was recently shown to be overexpressed in high-hyperdiploid pediatric ALLs with dup(1q), suggesting that gain of this gene may indeed underlie duplication of 1q, one of the most common secondary abnormalities in ALL (Table S4) (28). Two cases were assigned to different karyotypic subgroups based on the SNP array findings, including one iAMP21, a group that was recently reported to have an inferior outcome in pediatric ALL (21). Finally, two cases with t(9;22) displayed microdeletions adjacent to the breakpoints, showing that array analysis may, in some cases, also be used to identify rearrangements that are balanced on the cytogenetic level. Thus, SNP array analysis may give additional diagnostic and prognostic information in cases where G banding results are normal or not available.
In conclusion, by using high-resolution SNP arrays, we detect a very high frequency of hidden genetic changes in adult ALL cases. Deletions, commonly cryptic, comprised >70% of the found abnormalities, suggesting that microdeletions are a characteristic feature of adult ALL. Furthermore, the analyses revealed recurrent genetic abnormalities in adult ALL targeting genes not previously implicated in leukemogenesis. Most importantly, we report that the pattern of deletions seen in adult and adolescent ALL strongly resembles what has recently been shown for pediatric ALL, suggesting that microdeletions of key genes may be a general, characteristic feature of ALL that is shared among different clinical, morphological, and cytogenetic subgroups.
Materials and Methods
Patients.
Approximately 300 patients were diagnosed with adult ALL at St Bartholomew's Hospital, London, U.K., between 1980 and 2005. The present study included 45 (≈15%) cases, selected on the basis of available material from diagnosis for DNA extraction (Table S3). The median age at diagnosis was 33 years (13–69 years; Table S3); 14 patients were adolescents (21 years old or younger). Twenty-six cases were diagnosed with ALL of B cell lineage, 15 with T ALL, and four were ALL cases of unknown immunophenotype (Table S3). Diagnostic G banding and/or interphase FISH had been performed in 44 of the cases, showing that 10 had a t(9;22)(q34;q11), two harbored a t(4;11)(q21;q23) resulting in a MLL/AFF1 (previously AF4, MLLT2) fusion, and one had a t(8;14)(q24.1;q32) juxtaposing IGH and MYC. The remaining cases displayed various genetic abnormalities or were cytogenetically normal (Table S3). Presence of a p190 or p210 BCR/ABL1 fusion transcript was ascertained with standard RT-PCR in all t(9;22)-positive cases except case 9, for which RNA was unavailable.
SNP Array Analysis.
DNA was extracted according to standard methods from bone marrow or peripheral blood samples acquired at diagnosis. For SNP array analysis, the Affymetrix GeneChip Human Mapping 250K Nsp, 250K Sty, and 10K 2.0 arrays were used, together comprising ≈510,000 SNPs, with a median physical distance between SNPs of <2.5 kb (Affymetrix). SNP array analysis was performed according to the manufacturer's instructions (Affymetrix), except for post-PCR washes that were done with Ultrafree-MC columns (Millipore). GTYPE (Affymetrix) was used for analysis of signal intensity and for genotype calling. Raw data files are available from the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under series record GSE9611.
Data Analysis.
For copy number estimates and identification of regions with UPD, the in-house Genome Orientated Laboratory File (GOLF) software package was used (available online at http://bioinformatics.cancerresearchuk.org/cazier01/). Hybridization values were normalized to the median value on each array. Copy number was determined based on the log2 ratio of the signal intensity from the leukemia sample versus the pooled signal intensity of 10 unrelated nonleukemic control DNA samples and was done by visual inspection. Regions of UPD were defined as a maximum of two heterozygous calls in a window of 50 consecutive SNPs (29). Only putative copy-number changes involving at least three SNPs were included. Copy-number changes were also confirmed with dChipSNP by using Hidden Markov Model and/or visual inspection (30).
Statistical Analyses.
Possible associations between deletions of CDKN2A, PAX5, IKZF1, ETV6, or RB1 and clinical parameters were analyzed with the χ2 test, two-sided Fisher's exact test (gender, B or T lineage) and two-sided Mann–Whitney test (age, WCC) (VassarStats: Website for Statistical Computation; http://faculty.vassar.edu/lowry/VassarStats.html). The possible association between t(9;22) and IKZF1 deletion was analyzed with the Fisher's exact test. Comparisons between our results and published data from childhood ALL were performed by using the χ2 test and Fisher's exact test.
Supplementary Material
Acknowledgments.
We thank Drs. Michael Jenner and John Amess for diagnostic hematology and Dr. Jude Fitzgibbon, Ms. Amanda Dixon-McIver, Dr. Manu Gupta, and Dr. Manoj Raghavan for helpful discussion on the manuscript. This work was supported by grants from the Swedish Childhood Cancer Foundation, Cancer Research UK, and the Leukemia Research Fund.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE9611).
This article contains supporting information online at www.pnas.org/cgi/content/full/0800408105/DCSupplemental.
References
- 1.Faderl S, Jeha S, Kantarjian HM. The biology and therapy of adult acute lymphoblastic leukemia. Cancer. 2003;98:1337–1354. doi: 10.1002/cncr.11664. [DOI] [PubMed] [Google Scholar]
- 2.Rodriguez-Abreu D, Bordoni A, Zucca E. Epidemiology of hematological malignancies. Ann Oncol. 2007;18:i3–i8. doi: 10.1093/annonc/mdl443. [DOI] [PubMed] [Google Scholar]
- 3.Rowe JM, et al. Induction therapy for adults with acute lymphoblastic leukemia: Results of more than 1500 patients from the international ALL trial: MRC UKALL XII/ECOG E2993. Blood. 2005;106:3760–3767. doi: 10.1182/blood-2005-04-1623. [DOI] [PubMed] [Google Scholar]
- 4.Le Q-H, et al. Proportion of long-term event-free survivors and lifetime of adult patients not cured after a standard acute lymphoblastic leukemia therapeutic program: Adult acute lymphoblastic leukemia-94 trial. Cancer. 2007;109:2058–2067. doi: 10.1002/cncr.22632. [DOI] [PubMed] [Google Scholar]
- 5.Annino L, et al. Treatment of adult acute lymphoblastic leukemia (ALL): Long-term follow-up of the GIMEMA ALL 0288 randomized study. Blood. 2002;99:863–871. doi: 10.1182/blood.v99.3.863. [DOI] [PubMed] [Google Scholar]
- 6.Hann I, et al. Benefit of intensified treatment for all children with acute lymphoblastic leukaemia: Results from MRC UKALL XI and MRC ALL97 randomised trials. Leukemia. 2000;14:356–363. doi: 10.1038/sj.leu.2401704. [DOI] [PubMed] [Google Scholar]
- 7.Schultz KR, et al. Risk and response-based classification of childhood B-precursor acute lymphoblastic leukemia: a combined analysis of prognostic markers from the Pediatric Oncology Group (POG) and Children's Cancer Group (CCG) Blood. 2006;109:926–935. doi: 10.1182/blood-2006-01-024729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramanujachar R, Richards S, Hann I, Webb D. Adolescents with acute lymphoblastic leukaemia: Emerging from the shadow of paediatric and adult treatment protocols. Pediatr Blood Cancer. 2006;47:748–756. doi: 10.1002/pbc.20776. [DOI] [PubMed] [Google Scholar]
- 9.Mancini M, et al. A comprehensive genetic classification of adult acute lymphoblastic leukemia (ALL): Analysis of the GIMEMA 0496 protocol. Blood. 2005;105:3434–3441. doi: 10.1182/blood-2004-07-2922. [DOI] [PubMed] [Google Scholar]
- 10.Moorman AV, et al. Karyotype is an independent prognostic factor in adult acute lymphoblastic leukemia (ALL): Analysis of cytogenetic data from patients treated on the Medical Research Council (MRC) UKALLXII/Eastern Cooperative Oncology Group (ECOG) 2993 trial. Blood. 2007;109:3189–3197. doi: 10.1182/blood-2006-10-051912. [DOI] [PubMed] [Google Scholar]
- 11.Wetzler M, et al. Prospective karyotype analysis in adult acute lymphoblastic leukemia: The cancer and leukemia Group B experience. Blood. 1999;93:3983–3993. [PubMed] [Google Scholar]
- 12.Group Francais de Cytogénétique Hématologique. Cytogenetic abnormalities in adult acute lymphoblastic leukemia: Correlations with hematologic findings outcome A Collaborative Study of the Group Francais de Cytogénétique Hématologique. Blood. 1996;87:3135–3142. [PubMed] [Google Scholar]
- 13.Johansson B, Mertens F, Mitelman F. Clinical and biological importance of cytogenetic abnormalities in childhood and adult acute lymphoblastic leukemia. Ann Med. 2004;36:492–503. doi: 10.1080/07853890410018808. [DOI] [PubMed] [Google Scholar]
- 14.Pinkel D, Albertson DG. Array comparative genomic hybridization and its applications in cancer. Nat Genet. 2005;37:S11–S17. doi: 10.1038/ng1569. [DOI] [PubMed] [Google Scholar]
- 15.Strefford JC, et al. Genome complexity in acute lymphoblastic leukemia is revealed by array-based comparative genomic hybridization. Oncogene. 2007;26:4306–4318. doi: 10.1038/sj.onc.1210190. [DOI] [PubMed] [Google Scholar]
- 16.Mullighan CG, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446:758–764. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- 17.Kuiper RP, et al. High-resolution genomic profiling of childhood ALL reveals novel recurrent genetic lesions affecting pathways involved in lymphocyte differentiation and cell cycle progression. Leukemia. 2007;21:1258–1266. doi: 10.1038/sj.leu.2404691. [DOI] [PubMed] [Google Scholar]
- 18.Raghavan M, et al. Genome-wide single nucleotide polymorphism analysis reveals frequent partial uniparental disomy due to somatic recombination in acute myeloid leukemias. Cancer Res. 2005;65:375–378. [PubMed] [Google Scholar]
- 19.Paulsson K, et al. High-resolution genome-wide array-based comparative genome hybridization reveals cryptic chromosome changes in AML and MDS cases with trisomy 8 as the sole cytogenetic aberration. Leukemia. 2006;20:840–846. doi: 10.1038/sj.leu.2404145. [DOI] [PubMed] [Google Scholar]
- 20.Iafrate AJ, et al. Detection of large-scale variation in the human genome. Nat Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 21.Moorman AV, et al. Prognosis of children with acute lymphoblastic leukemia (ALL) and intrachromosomal amplification of chromosome 21 (iAMP21) Blood. 2007;109:2327–2330. doi: 10.1182/blood-2006-08-040436. [DOI] [PubMed] [Google Scholar]
- 22.Roman-Gomez J, et al. Promoter hypermethylation of cancer-related genes: A strong independent prognostic factor in acute lymphoblastic leukemia. Blood. 2004;104:2492–2498. doi: 10.1182/blood-2004-03-0954. [DOI] [PubMed] [Google Scholar]
- 23.Faderl S, et al. The prognostic significance of p16INK4a/p14ARF locus deletion and MDM-2 protein expression in adult acute myelogenous leukemia. Cancer. 2000;89:1976–1982. doi: 10.1002/1097-0142(20001101)89:9<1976::aid-cncr14>3.3.co;2-e. [DOI] [PubMed] [Google Scholar]
- 24.Lee DS, et al. The presence of TEL/AML1 rearrangement and cryptic deletion of the TEL gene in adult acute lymphoblastic leukemia (ALL) Cancer Genet Cytogenet. 2005;162:176–178. doi: 10.1016/j.cancergencyto.2005.02.020. [DOI] [PubMed] [Google Scholar]
- 25.Kawamata N, et al. Molecular allelokaryotyping of pediatric acute lymphoblastic leukemias by high resolution single nucleotide polymorphism oligonucleotide genomic microarray. Blood. 2008;111:776–784. doi: 10.1182/blood-2007-05-088310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holmes ML, Pridans C, Nutt SL. The regulation of the B-cell gene expression programme by Pax5. Immunol Cell Biol. 2008;86:47–53. doi: 10.1038/sj.icb.7100134. [DOI] [PubMed] [Google Scholar]
- 27.Johansson B, Mertens F, Mitelman F. Primary vs. secondary neoplasia-associated chromosomal abnormalities—balanced rearrangements vs. genomic imbalances? Genes Chromosomes Cancer. 1996;16:155–163. doi: 10.1002/(SICI)1098-2264(199607)16:3<155::AID-GCC1>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 28.Davidsson J, et al. Tiling resolution array comparative genomic hybridization, expression and methylation analyses of dup(1q) in Burkitt lymphomas and pediatric high hyperdiploid acute lymphoblastic leukemias reveal clustered near-centromeric breakpoints and overexpression of genes in 1q22–32.3. Hum Mol Genet. 2007;16:2215–2225. doi: 10.1093/hmg/ddm173. [DOI] [PubMed] [Google Scholar]
- 29.Gupta, et al. Novel regions of acquired uniparental disomy discovered in acute myeloid leukemia. Genes Chromosomes Cancer. 2008 doi: 10.1002/gcc.20573. in press. [DOI] [PubMed] [Google Scholar]
- 30.Lin M, et al. dChipSNP: Significance curve and clustering of SNP-array-based loss-of-heterozygosity data. Bioinformatics. 2004;20:1233–1240. doi: 10.1093/bioinformatics/bth069. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.