Abstract
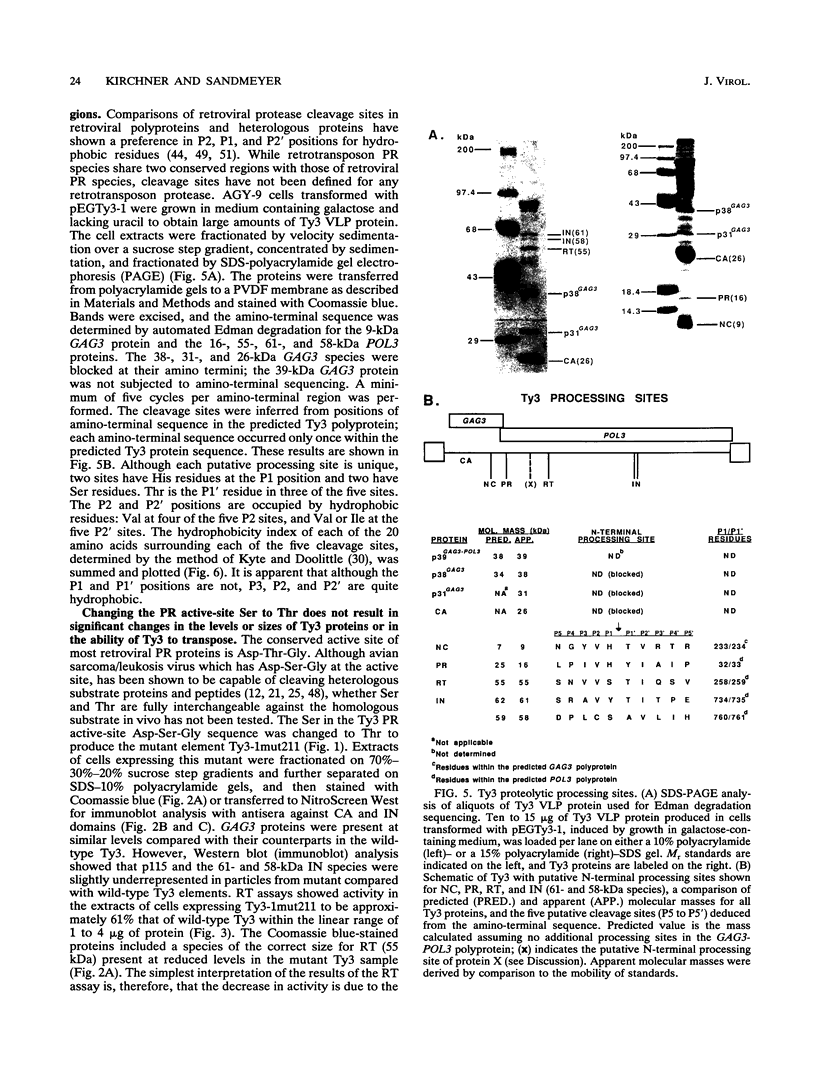
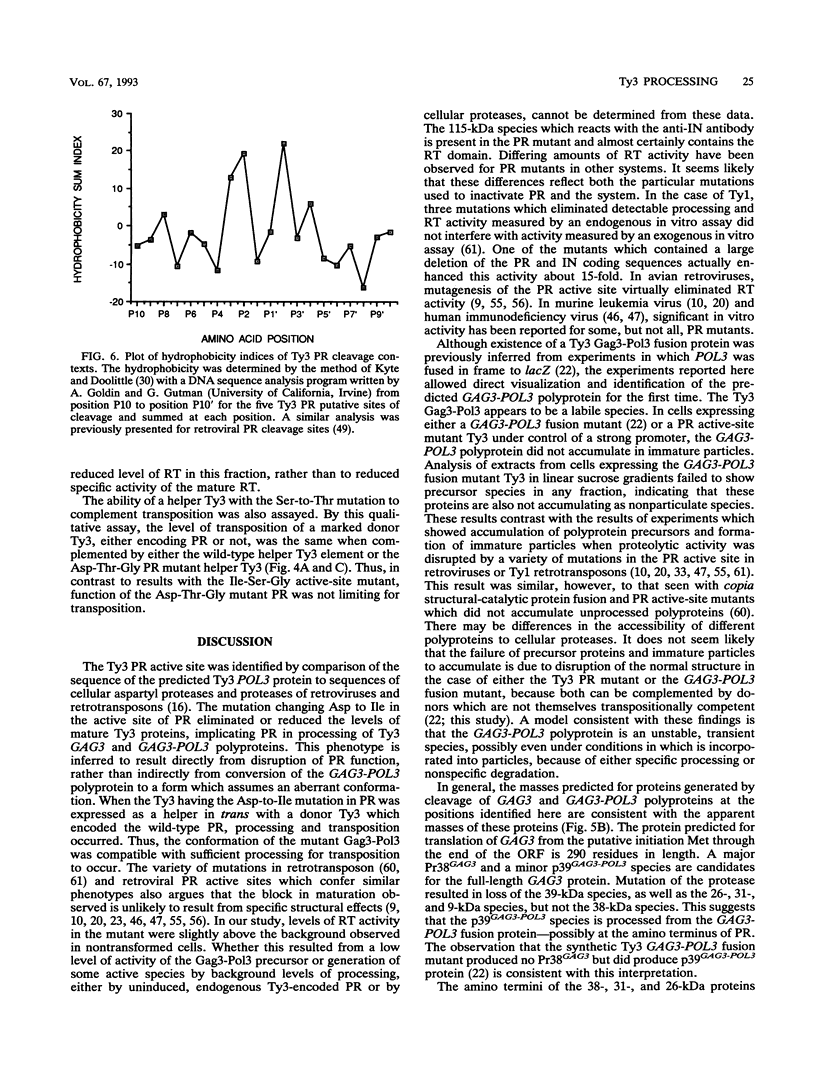
Ty3 is a retroviruslike element found in Saccharomyces cerevisiae. It encodes GAG3 and GAG3-POL3 polyproteins which are processed into mature proteins found in the Ty3 viruslike particle. In this study, the region encoding a protease that is homologous to retroviral aspartyl proteases was identified and shown to be required for production of mature Ty3 proteins and transposition. The Ty3 protease has the Asp-Ser-Gly consensus sequence found in copia, Ty1, and Rous sarcoma virus proteases, rather than the Asp-Thr-Gly found in most retroviral proteases. The Asp-Ser-Gly consensus is flanked by residues similar to those which flank the active sites of cellular aspartyl proteases. Mutations were made in the Ty3 active-site sequence to examine the role of the protease in Ty3 particle maturation and to test the functional significance of the Ser active-site variant in the consensus sequence. Mutation of the active-site Asp blocked processing of Gag3 and Gag3-Pol3 and allowed identification of a GAG3-POL3 polyprotein. This protein was turned over rapidly in cells expressing the mutant Ty3. Changing the active-site Ser to Thr caused only a modest reduction in the levels of certain Ty3 proteins. Five putative cleavage sites of this protease in Ty3 GAG3 and GAG3-POL3 polyproteins were defined by amino-terminal sequence analysis. The existence of an additional protein(s) of unknown function, encoded downstream of the protease-coding region, was deduced from the positions of these amino termini and the sizes of known Ty3 proteins. Although Ty3 protease cleavage sites do not correspond exactly to known retroviral protease cleavage sites, there are similarities. Residues P3 through P2' in the regions encompassing each of the five sites are uncharged, and no P1 position is occupied by an amino acid with a branched beta carbon.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Burstein H., Bizub D., Skalka A. M. Assembly and processing of avian retroviral gag polyproteins containing linked protease dimers. J Virol. 1991 Nov;65(11):6165–6172. doi: 10.1128/jvi.65.11.6165-6172.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalker D. L., Sandmeyer S. B. Transfer RNA genes are genomic targets for de Novo transposition of the yeast retrotransposon Ty3. Genetics. 1990 Dec;126(4):837–850. doi: 10.1093/genetics/126.4.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark D. J., Bilanchone V. W., Haywood L. J., Dildine S. L., Sandmeyer S. B. A yeast sigma composite element, TY3, has properties of a retrotransposon. J Biol Chem. 1988 Jan 25;263(3):1413–1423. [PubMed] [Google Scholar]
- Craven R. C., Bennett R. P., Wills J. W. Role of the avian retroviral protease in the activation of reverse transcriptase during virion assembly. J Virol. 1991 Nov;65(11):6205–6217. doi: 10.1128/jvi.65.11.6205-6217.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford S., Goff S. P. A deletion mutation in the 5' part of the pol gene of Moloney murine leukemia virus blocks proteolytic processing of the gag and pol polyproteins. J Virol. 1985 Mar;53(3):899–907. doi: 10.1128/jvi.53.3.899-907.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debouck C., Gorniak J. G., Strickler J. E., Meek T. D., Metcalf B. W., Rosenberg M. Human immunodeficiency virus protease expressed in Escherichia coli exhibits autoprocessing and specific maturation of the gag precursor. Proc Natl Acad Sci U S A. 1987 Dec;84(24):8903–8906. doi: 10.1073/pnas.84.24.8903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demsey A., Collins F., Kawka D. Structure of and alterations to defective murine sarcoma virus particles lacking envelope proteins and core polyprotein cleavage. J Virol. 1980 Dec;36(3):872–877. doi: 10.1128/jvi.36.3.872-877.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichinger D. J., Boeke J. D. The DNA intermediate in yeast Ty1 element transposition copurifies with virus-like particles: cell-free Ty1 transposition. Cell. 1988 Sep 23;54(7):955–966. doi: 10.1016/0092-8674(88)90110-9. [DOI] [PubMed] [Google Scholar]
- Garfinkel D. J., Hedge A. M., Youngren S. D., Copeland T. D. Proteolytic processing of pol-TYB proteins from the yeast retrotransposon Ty1. J Virol. 1991 Sep;65(9):4573–4581. doi: 10.1128/jvi.65.9.4573-4581.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen L. J., Chalker D. L., Orlinsky K. J., Sandmeyer S. B. Ty3 GAG3 and POL3 genes encode the components of intracellular particles. J Virol. 1992 Mar;66(3):1414–1424. doi: 10.1128/jvi.66.3.1414-1424.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen L. J., Chalker D. L., Sandmeyer S. B. Ty3, a yeast retrotransposon associated with tRNA genes, has homology to animal retroviruses. Mol Cell Biol. 1988 Dec;8(12):5245–5256. doi: 10.1128/mcb.8.12.5245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen L. J., Sandmeyer S. B. Characterization of a transpositionally active Ty3 element and identification of the Ty3 integrase protein. J Virol. 1990 Jun;64(6):2599–2607. doi: 10.1128/jvi.64.6.2599-2607.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Elliott R. C., Liu P. S., Koduri R. K., Weickmann J. L., Lee J. H., Blair L. C., Ghosh-Dastidar P., Bradshaw R. A., Bryan K. M. Specificity of cotranslational amino-terminal processing of proteins in yeast. Biochemistry. 1987 Dec 15;26(25):8242–8246. doi: 10.1021/bi00399a033. [DOI] [PubMed] [Google Scholar]
- Katoh I., Ikawa Y., Yoshinaka Y. Retrovirus protease characterized as a dimeric aspartic proteinase. J Virol. 1989 May;63(5):2226–2232. doi: 10.1128/jvi.63.5.2226-2232.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh I., Yoshinaka Y., Rein A., Shibuya M., Odaka T., Oroszlan S. Murine leukemia virus maturation: protease region required for conversion from "immature" to "mature" core form and for virus infectivity. Virology. 1985 Sep;145(2):280–292. doi: 10.1016/0042-6822(85)90161-8. [DOI] [PubMed] [Google Scholar]
- Khan A. S., Stephenson J. R. Feline sarcoma virus-coded polyprotein: enzymatic cleavage by a type C virus-coded structural protein. J Virol. 1979 Feb;29(2):649–656. doi: 10.1128/jvi.29.2.649-656.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchner J., Sandmeyer S. B., Forrest D. B. Transposition of a Ty3 GAG3-POL3 fusion mutant is limited by availability of capsid protein. J Virol. 1992 Oct;66(10):6081–6092. doi: 10.1128/jvi.66.10.6081-6092.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohl N. E., Emini E. A., Schleif W. A., Davis L. J., Heimbach J. C., Dixon R. A., Scolnick E. M., Sigal I. S. Active human immunodeficiency virus protease is required for viral infectivity. Proc Natl Acad Sci U S A. 1988 Jul;85(13):4686–4690. doi: 10.1073/pnas.85.13.4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotler M., Danho W., Katz R. A., Leis J., Skalka A. M. Avian retroviral protease and cellular aspartic proteases are distinguished by activities on peptide substrates. J Biol Chem. 1989 Feb 25;264(6):3428–3435. [PubMed] [Google Scholar]
- Kotler M., Katz R. A., Danho W., Leis J., Skalka A. M. Synthetic peptides as substrates and inhibitors of a retroviral protease. Proc Natl Acad Sci U S A. 1988 Jun;85(12):4185–4189. doi: 10.1073/pnas.85.12.4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotler M., Katz R. A., Skalka A. M. Activity of avian retroviral protease expressed in Escherichia coli. J Virol. 1988 Aug;62(8):2696–2700. doi: 10.1128/jvi.62.8.2696-2700.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer R. A., Schaber M. D., Skalka A. M., Ganguly K., Wong-Staal F., Reddy E. P. HTLV-III gag protein is processed in yeast cells by the virus pol-protease. Science. 1986 Mar 28;231(4745):1580–1584. doi: 10.1126/science.2420008. [DOI] [PubMed] [Google Scholar]
- Kräusslich H. G. Human immunodeficiency virus proteinase dimer as component of the viral polyprotein prevents particle assembly and viral infectivity. Proc Natl Acad Sci U S A. 1991 Apr 15;88(8):3213–3217. doi: 10.1073/pnas.88.8.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Le Grice S. F., Mills J., Mous J. Active site mutagenesis of the AIDS virus protease and its alleviation by trans complementation. EMBO J. 1988 Aug;7(8):2547–2553. doi: 10.1002/j.1460-2075.1988.tb03103.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee F. J., Lin L. W., Smith J. A. Model peptides reveal specificity of N alpha-acetyltransferase from Saccharomyces cerevisiae. J Biol Chem. 1990 Jul 15;265(20):11576–11580. [PubMed] [Google Scholar]
- Loeb D. D., Hutchison C. A., 3rd, Edgell M. H., Farmerie W. G., Swanstrom R. Mutational analysis of human immunodeficiency virus type 1 protease suggests functional homology with aspartic proteinases. J Virol. 1989 Jan;63(1):111–121. doi: 10.1128/jvi.63.1.111-121.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb D. D., Swanstrom R., Everitt L., Manchester M., Stamper S. E., Hutchison C. A., 3rd Complete mutagenesis of the HIV-1 protease. Nature. 1989 Aug 3;340(6232):397–400. doi: 10.1038/340397a0. [DOI] [PubMed] [Google Scholar]
- McClure M. A., Johnson M. S., Doolittle R. F. Relocation of a protease-like gene segment between two retroviruses. Proc Natl Acad Sci U S A. 1987 May;84(9):2693–2697. doi: 10.1073/pnas.84.9.2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meek T. D., Dayton B. D., Metcalf B. W., Dreyer G. B., Strickler J. E., Gorniak J. G., Rosenberg M., Moore M. L., Magaard V. W., Debouck C. Human immunodeficiency virus 1 protease expressed in Escherichia coli behaves as a dimeric aspartic protease. Proc Natl Acad Sci U S A. 1989 Mar;86(6):1841–1845. doi: 10.1073/pnas.86.6.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M., Jaskólski M., Rao J. K., Leis J., Wlodawer A. Crystal structure of a retroviral protease proves relationship to aspartic protease family. Nature. 1989 Feb 9;337(6207):576–579. doi: 10.1038/337576a0. [DOI] [PubMed] [Google Scholar]
- Navia M. A., Fitzgerald P. M., McKeever B. M., Leu C. T., Heimbach J. C., Herber W. K., Sigal I. S., Darke P. L., Springer J. P. Three-dimensional structure of aspartyl protease from human immunodeficiency virus HIV-1. Nature. 1989 Feb 16;337(6208):615–620. doi: 10.1038/337615a0. [DOI] [PubMed] [Google Scholar]
- Oroszlan S., Luftig R. B. Retroviral proteinases. Curr Top Microbiol Immunol. 1990;157:153–185. doi: 10.1007/978-3-642-75218-6_6. [DOI] [PubMed] [Google Scholar]
- Palmiter R. D., Gagnon J., Vogt V. M., Ripley S., Eisenman R. N. The NH2-terminal sequence of the avian oncovirus gag precursor polyprotein (Pr76gag). Virology. 1978 Dec;91(2):423–433. doi: 10.1016/0042-6822(78)90388-4. [DOI] [PubMed] [Google Scholar]
- Pearl L. H., Taylor W. R. A structural model for the retroviral proteases. Nature. 1987 Sep 24;329(6137):351–354. doi: 10.1038/329351a0. [DOI] [PubMed] [Google Scholar]
- Pearl L. H., Taylor W. R. Sequence specificity of retroviral proteases. Nature. 1987 Aug 6;328(6130):482–482. doi: 10.1038/328482b0. [DOI] [PubMed] [Google Scholar]
- Peng C., Chang N. T., Chang T. W. Identification and characterization of human immunodeficiency virus type 1 gag-pol fusion protein in transfected mammalian cells. J Virol. 1991 May;65(5):2751–2756. doi: 10.1128/jvi.65.5.2751-2756.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng C., Ho B. K., Chang T. W., Chang N. T. Role of human immunodeficiency virus type 1-specific protease in core protein maturation and viral infectivity. J Virol. 1989 Jun;63(6):2550–2556. doi: 10.1128/jvi.63.6.2550-2556.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepinsky R. B. Localization of lipid-protein and protein-protein interactions within the murine retrovirus gag precursor by a novel peptide-mapping technique. J Biol Chem. 1983 Sep 25;258(18):11229–11235. [PubMed] [Google Scholar]
- Pettit S. C., Simsic J., Loeb D. D., Everitt L., Hutchison C. A., 3rd, Swanstrom R. Analysis of retroviral protease cleavage sites reveals two types of cleavage sites and the structural requirements of the P1 amino acid. J Biol Chem. 1991 Aug 5;266(22):14539–14547. [PubMed] [Google Scholar]
- Poorman R. A., Tomasselli A. G., Heinrikson R. L., Kézdy F. J. A cumulative specificity model for proteases from human immunodeficiency virus types 1 and 2, inferred from statistical analysis of an extended substrate data base. J Biol Chem. 1991 Aug 5;266(22):14554–14561. [PubMed] [Google Scholar]
- Power M. D., Marx P. A., Bryant M. L., Gardner M. B., Barr P. J., Luciw P. A. Nucleotide sequence of SRV-1, a type D simian acquired immune deficiency syndrome retrovirus. Science. 1986 Mar 28;231(4745):1567–1572. doi: 10.1126/science.3006247. [DOI] [PubMed] [Google Scholar]
- Skalka A. M. Retroviral proteases: first glimpses at the anatomy of a processing machine. Cell. 1989 Mar 24;56(6):911–913. doi: 10.1016/0092-8674(89)90621-1. [DOI] [PubMed] [Google Scholar]
- Staden R. Methods to define and locate patterns of motifs in sequences. Comput Appl Biosci. 1988 Mar;4(1):53–60. doi: 10.1093/bioinformatics/4.1.53. [DOI] [PubMed] [Google Scholar]
- Stewart L., Schatz G., Vogt V. M. Properties of avian retrovirus particles defective in viral protease. J Virol. 1990 Oct;64(10):5076–5092. doi: 10.1128/jvi.64.10.5076-5092.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart L., Vogt V. M. trans-acting viral protease is necessary and sufficient for activation of avian leukosis virus reverse transcriptase. J Virol. 1991 Nov;65(11):6218–6231. doi: 10.1128/jvi.65.11.6218-6231.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh H., Kikuno R., Hayashida H., Miyata T., Kugimiya W., Inouye S., Yuki S., Saigo K. Close structural resemblance between putative polymerase of a Drosophila transposable genetic element 17.6 and pol gene product of Moloney murine leukaemia virus. EMBO J. 1985 May;4(5):1267–1272. doi: 10.1002/j.1460-2075.1985.tb03771.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tritch R. J., Cheng Y. E., Yin F. H., Erickson-Viitanen S. Mutagenesis of protease cleavage sites in the human immunodeficiency virus type 1 gag polyprotein. J Virol. 1991 Feb;65(2):922–930. doi: 10.1128/jvi.65.2.922-930.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wlodawer A., Miller M., Jaskólski M., Sathyanarayana B. K., Baldwin E., Weber I. T., Selk L. M., Clawson L., Schneider J., Kent S. B. Conserved folding in retroviral proteases: crystal structure of a synthetic HIV-1 protease. Science. 1989 Aug 11;245(4918):616–621. doi: 10.1126/science.2548279. [DOI] [PubMed] [Google Scholar]
- Yoshioka K., Honma H., Zushi M., Kondo S., Togashi S., Miyake T., Shiba T. Virus-like particle formation of Drosophila copia through autocatalytic processing. EMBO J. 1990 Feb;9(2):535–541. doi: 10.1002/j.1460-2075.1990.tb08140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngren S. D., Boeke J. D., Sanders N. J., Garfinkel D. J. Functional organization of the retrotransposon Ty from Saccharomyces cerevisiae: Ty protease is required for transposition. Mol Cell Biol. 1988 Apr;8(4):1421–1431. doi: 10.1128/mcb.8.4.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]