Abstract
In Escherichia coli, the glutathione/glutaredoxin and thioredoxin pathways are essential for the reduction of cytoplasmic protein disulfide bonds, including those formed in the essential enzyme ribonucleotide reductase during its action on substrates. Double mutants lacking thioredoxin reductase (trxB) and glutathione reductase (gor) or glutathione biosynthesis (gshA) cannot grow. Growth of Δgor ΔtrxB strains is restored by a mutant (ahpC*) of the peroxiredoxin AhpC, converting it to a disulfide reductase that generates reduced glutathione. Here, we show that ahpC* also restores growth to a ΔgshB ΔtrxB strain, which lacks glutathione and accumulates only its precursor γ-glutamylcysteine (γ-GC). It suppresses this strain by allowing accumulation of reduced γ-GC, which can substitute for glutathione. Surprisingly, new ahpC suppressor mutations arose in a ΔgshA ΔtrxB strain lacking both glutathione and γ-GC, a strain that ahpC* does not suppress. Some of these mutant AhpC proteins channel electrons into the disulfide-reducing pathways via either the thioredoxins or the glutaredoxins without, evidently, the intermediary of glutathione. Our results provide insights into the physiological functioning of the glutathione pathway and reveal surprising plasticity of a peroxidase because different mutant versions of AhpC can channel electrons into the disulfide-reducing pathways by at least four distinct routes. Despite the reductase activity of mutant AhpCs, these various suppressor strains exhibit an oxidizing cytoplasm and accumulate correctly folded disulfide-bonded proteins in their cytoplasm. Proteins most effectively oxidized vary between strains, potentially providing useful tools for expressing different disulfide-bonded proteins.
Keywords: cytoplasmic oxidative folding, disulfide bonds, glutathione, peroxiredoxin (ahpC), suppressor mutations
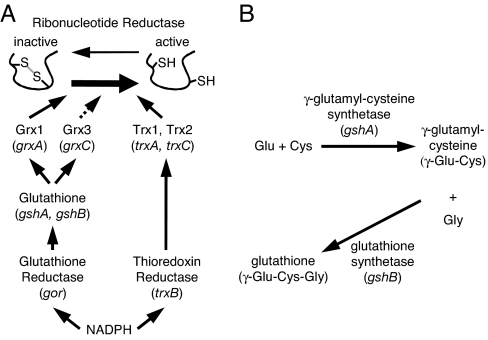
The control over whether disulfide bonds accumulate in cytoplasmic proteins in Escherichia coli is determined by two reductive pathways. The thioredoxin/thioredoxin reductase and glutathione/glutaredoxin pathways use electrons derived from NADPH to maintain active-site cysteines of enzymes such as ribonucleotide reductase in the reduced state and, under oxidative stress conditions, prevent the formation of aberrant disulfide bonds in proteins (Fig. 1A). Both of these pathways contain small, 9- to 25-kDa proteins belonging to the thioredoxin superfamily, either glutaredoxins or thioredoxins. This protein family contains the consensus motif CXXC necessary for thiol redox activity, whether it is oxidation or reduction.
Fig. 1.
Cytoplasmic thiol redox pathway components. (A) Cytoplasmic thiol redox pathways of E. coli. The dashed line for Grx3 indicates that it can reduce ribonucleotide reductase but does so inefficiently. Not shown are glutaredoxin 2 and glutaredoxin 4, which are unable to reduce ribonucleotide reductase. Grx, glutaredoxin; Trx, thioredoxin. (B) Glutathione biosynthesis pathway.
E. coli is not viable when both reductive pathways are disrupted by deletion of the enzymes glutathione reductase (gor) and thioredoxin reductase (trxB). This lack of viability is likely to caused by the inability to reduce ribonucleotide reductase (1, 2). However, extragenic suppressors can restore the viability of a Δgor ΔtrxB strain (3). The suppressor mutations all map to the gene ahpC, which encodes a peroxiredoxin whose physiological function is the reduction of hydrogen peroxide and alkylhydroperoxides. The ahpC suppressor mutations, including a triplet repeat expansion (ahpC*) (3), encode AhpC proteins that show greatly enhanced activity toward the reduction of glutathionylated glutaredoxins (4). This reaction yields reduced glutathione, which can mediate the recycling of ribonucleotide reductase and presumably of other cytoplasmic enzymes that employ cysteine redox chemistry in their catalytic cycles.
The above results underscore the key role of glutathione (GSH) in cellular adaptation to the inactivation of the protein reduction pathways. They raise the fundamental questions of whether, or how, the cell can respond to the simultaneous absence of GSH and reduced thioredoxins. The answer to this question has important mechanistic and practical ramifications. First, the isolation of mutants that can grow despite the absence of GSH and reduced thioredoxins would indicate the existence of mechanisms for the reduction of ribonucleotide reductase. Second, the absence of the physiological reductants GSH and thioredoxin would likely affect the kinetics of formation of protein disulfide bonds in the cytoplasm in a manner that is distinct from that of the Δgor ΔtrxB ahpC* strain that still contains a GSH redox buffer. Third, new suppressor strains could provide an optimal oxidative environment for the folding of heterologous proteins that are difficult to produce in Origami, a Δgor ΔtrxB ahpC* strain (5).
In E. coli, GSH is synthesized by the enzymes γ-glutamylcysteine synthetase (gshA) and glutathione synthetase (gshB). The first enzyme produces γ-glutamylcysteine (γ-GC), which glutathione synthetase then conjugates to glycine, yielding GSH (Fig. 1B). Here, we show that the ΔgshA ΔtrxB and ΔgshB ΔtrxB strains exhibit growth defects, and we report on the isolation and characterization of suppressor mutations that restore growth of the resulting strains. To our surprise, most suppressor mutations of the ΔgshA ΔtrxB strain and all suppressor mutations of the ΔgshB ΔtrxB strain were in the ahpC gene. Additionally, these mutant ahpCs appear to mediate suppression in mechanistically distinct ways. For the new mutant ahpCs isolated in the ΔgshA ΔtrxB supp strains, suppression occurs in the absence of GSH and γ-GC, the biosynthetic precursor to GSH. This suppression contrasts with suppression by mutant ahpCs previously isolated (3, 4), which we show require at least γ-GC. These findings underscore the functional plasticity of alkyl hydroperoxidase and show that plasticity is able to provide a means of adapting to an oxidizing cytoplasm. Furthermore, the ΔgshB ΔtrxB supp and ΔgshA ΔtrxB supp strains displayed a capacity to make disulfide bonds in normally secreted proteins when removal of their signal sequences forced their retention in the cytoplasm. However, the different suppressor strains favor disulfide bond formation in different ones of these cytoplasmically localized substrate proteins, indicating that this collection of strains can be useful more generally for the expression of a variety of such proteins.
Results
Construction and Properties of ΔgshA ΔtrxB and ΔgshB ΔtrxB Strains.
We set out to obtain new classes of suppressors of mutants defective in cytoplasmic reducing pathways. Because previous suppressor mutations from the Δgor ΔtrxB strain, which were all in ahpC, were dependent on gshA for their activity, we constructed strains with deletions in both the GSH biosynthesis and the thioredoxin reduction pathways (ΔgshA ΔtrxB and ΔgshB ΔtrxB). We constructed these strains in the presence of a high-copy-complementing plasmid (pMJF1 or pMJF2), which confers ampicillin resistance and expresses either the gshA or the gshB gene, respectively, under the tightly controlled PBAD promoter. In the absence of the inducer arabinose, the ΔgshB ΔtrxB strain forms small colonies after 2 days, whereas the ΔgshA ΔtrxB strain does not form colonies. The parental strain DHB4 forms large colonies within 1 day.
Earlier we had found that the addition of the reductant DTT allows growth of Δgor ΔtrxB strains (6). Here, we found that, in the absence of arabinose, ΔgshA ΔtrxB and ΔgshB ΔtrxB strains containing vectors pMJF1 or pMJF2 also grew well in media containing DTT (50 μl of 1 M DTT on a filter disk). Based on this finding and to eliminate complications caused by the complementing plasmid, we obtained derivatives of the two double-mutant strains cured of their complementing plasmids, growing strains in the absence of ampicillin and in the presence of DTT. We readily detected cells that had lost their ampicillin resistance as a result of loss of the plasmid. The ΔgshA ΔtrxB and ΔgshB ΔtrxB strains without plasmid formed good-sized colonies after 1 day on rich agar media containing DTT. Whereas in the absence of DTT the ΔgshA ΔtrxB strain failed to grow at all, the ΔgshB ΔtrxB strain did grow, albeit slowly.
Suppressors of the ΔgshB ΔtrxB Strain Are ahpC* Mutations.
We first selected for suppressors of the ΔgshB ΔtrxB strain, growing it in the presence of DTT and then plating cells on rich media lacking DTT. Suppressor mutants, which formed large colonies within 1 day, arose at a frequency of ≈10−4. Nine suppressor strains were selected for further analysis. Suppression depended on the presence of glutaredoxin 1 and the first enzyme of GSH biosynthesis, γ-GC synthetase; P1 transduction of null alleles of either grxA or gshA into the suppressed strains eliminated suppressor activity. Furthermore, and to our surprise, suppression also required the presence of the ahpCF locus, indicating that suppression might be caused by ahpC mutations.
Sequencing the ahpC gene in four of the ΔgshB ΔtrxB supp strains showed the same triplet repeat expansion mutation, ahpC*, originally found as a suppressor of the Δgor ΔtrxB strain. We confirmed that ahpC* was sufficient to suppress the ΔgshB ΔtrxB mutant by showing that P1 transduction of the original ahpC* isolate with a linked Tn10 transposon (3) into a ΔgshB ΔtrxB strain restored normal growth in the absence of DTT. Whereas ahpC* effectively suppresses both Δgor ΔtrxB and ΔgshB ΔtrxB strains, the latter suppressed strain grew somewhat more slowly than the former in both rich and minimal media supplemented with amino acids.
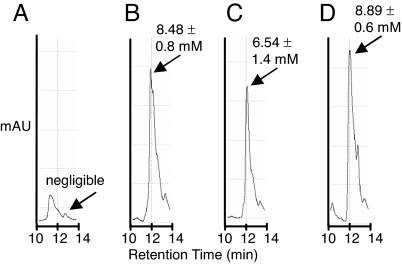
The ahpC* mutation is a gain-of-function mutation that enables the alkyl hydroperoxidase enzyme to reduce glutathionylated glutaredoxins, thus generating reduced GSH (4, 7). Thus, in the Δgor ΔtrxB ahpC* strains, the reductase activity of AhpC* maintains the reduced GSH pool at a level essentially indistinguishable from that found in the wild-type parental strain [supporting information (SI) Fig. S1A]. Analogously, we examined the mutant strain ΔgshB ΔtrxB ahpC* to determine the level of γ-GC and its redox state. Stationary phase cells were incubated with the thiol-specific reagent monobromobimane (mBBr), lysed, proteins were precipitated with trichloroacetic acid, and the concentration of γ-GC-mBBr was determined by HPLC. LC-MS revealed only one peak with a 12-min retention time and m/z = 441 corresponding to γ-GC-mBBr (Fig. S1C). The level of total (reduced + oxidized) γ-GC was determined by first reducing cell lysates with Tris(2-carboxyethyl) phosphine (TCEP) before derivatization with mBBr. Of a total of 8.89 ± 0.6 mM γ-GC, 6.54 ± 1.4 mM were reduced (Fig. 2 C and D). Thus, ≈75% of the γ-GC is present in the reduced state in the ΔgshB ΔtrxB ahpC* strain. Conversely, γ-GC was predominantly oxidized in the parental ΔgshB ΔtrxB strain containing wild-type ahpC (Fig. 2 A and B).
Fig. 2.
Reverse-phase HPLC of γ-GC-mBBr derivatives. Values correspond to the intracellular concentration of γ-GC. Errors were calculated from two independent experiments. (A) Reduced γ-GC in the ΔgshB ΔtrxB strain. (B) Total γ-GC in the ΔgshB ΔtrxB strain. (C) Reduced γ-GC in the ΔgshB ΔtrxB ahpC* strain. (D) Total γ-GC in the ΔgshB ΔtrxB ahpC* strain.
A recent extensive search for suppressor alleles of the Δgor ΔtrxB strain yielded new suppressor mutations that cause single amino acid substitutions in AhpC: C46Y, R119C, S159P, P161S, P166L, and A167T. Like AhpC*, these newly characterized mutant enzymes exhibited enhanced ability to reduce glutathionylated glutaredoxins (4). We find that these mutant ahpC alleles also suppressed the growth defect of the ΔgshB ΔtrxB strain when expressed from a plasmid or transduced into the strain with a linked Tn10 transposon (Table 1). The ΔgshB ΔtrxB strain containing these ahpC suppressor alleles grew slightly more slowly than the respective Δgor ΔtrxB strain in rich media.
Table 1.
Suppression of mutants of the cytoplasmic redox pathways by mutant AhpC
| Change in AhpC | Growth of strain on NZ† |
|||
|---|---|---|---|---|
| DHB4 | Δgor ΔtrxB | ΔgshB ΔtrxB | ΔgshA ΔtrxB | |
| Wild type | +++ | − | Very tiny | − |
| Isolated as suppressors of Δgor ΔtrxB | ||||
| AhpC* (+F38) | +++ | +++ | +++ | − |
| C46Y | +++ | +++ | ++ | − |
| R119C | +++ | +++ | ++ | − |
| G141S | +++ | ++ | ++ | − |
| S159P | +++ | +++ | +++ | − |
| P161S | +++ | ++ | + | − |
| P166S | +++ | +++ | +++ | − |
| P166L | +++ | +++ | +++ | − |
| A167T | +++ | +++ | +++ | − |
| Isolated as suppressors of ΔgshA ΔtrxB | ||||
| S71F | +++ | +++ | ++ | + |
| S71F, ΔE173 | +++ | +++ | ++ | ++ |
| V164G | +++ | +++ | +++ | ++ |
| G162–W169 | ND | ND‡ | ND | ++ |
| E171Ter | +++ | ++ | ++ | ++ |
†+++, grows similar to wild type (DHB4); ++, grows well but forms smaller colonies than wild type; +, forms tiny colonies after 1 day; Very tiny, forms good-sized colonies only after 2 days; −, does not grow; ND, not determined.
‡Deletion of the gene gor in the Δ gshA Δ trxB ahpC (G162–W169) strain does not result in loss of suppression.
A Different Class of ahpC Mutations Is Required for Suppression of the ΔgshA ΔtrxB Strain.
In contrast to the results with the ΔgshB ΔtrxB strain, the growth defect of the ΔgshA ΔtrxB strain was not suppressed by the ahpC* mutation nor by the other ahpC suppressor mutations (Table 1). Therefore, to isolate new classes of suppressor mutations, we grew the ΔgshA ΔtrxB strain in the presence of DTT and plated it on rich media without DTT. Colonies arose at a frequency of 10−7 after a 2-day incubation, a frequency ≈3 orders of magnitude lower than that found with the ΔgshB ΔtrxB or Δgor ΔtrxB strains. The ΔgshA ΔtrxB suppressor strains also grew more slowly than the ΔgshB ΔtrxB or Δgor ΔtrxB suppressor strains, forming only small colonies after 1 day on rich media at 37°C.
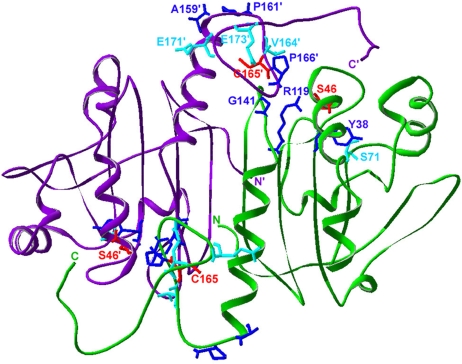
We selected 10 ΔgshA ΔtrxB suppressor strains to analyze. Surprisingly, P1 transduction mapping with a Tn10 transposon linked to ahpC indicated that all suppressors were linked to ahpCF. DNA sequencing of ahpC revealed the presence of mutations that altered the amino acid sequence of the AhpC protein in 6 of the 10 strains. Two isolates encoded a V164G amino acid substitution, one a S71F amino acid substitution, and one included S71F but also contained a deletion of the codon for E173. (The E173/S71F double mutant gives better suppression than S71F by itself; the E173 deletion by itself shows weaker suppression.) One clone contained a stop codon at position 171 (E171Ter), and the sixth suppressor strain contained a duplication of residues 162–169 (GEVCPAKW) of AhpC. Thus, most of the suppressor mutations alter residues close to one of the AhpC redox-active cysteines, C165 (Fig. 3).
Fig. 3.
Location of the residues found changed in AhpC in cytoplasmic thiol redox suppressor strains. Shown is an AhpC dimer (subunits in green and purple) in the structure of the C46S mutant (Protein Data Bank ID code 1N8J). C165 and C46 (replaced with serine in this structure), are shown in red. Residues found mutated in the ΔtrxB Δgor supp strain are in blue (3, 4). The ahpC* mutation results in the addition of a phenylalanine residue at Y38. Residues found mutated in the ΔgshA ΔtrxB supp strain are in cyan.
All six suppressor strains containing ahpC mutations grew on minimal media supplemented with glucose and amino acids but lacking GSH. We determined the concentration of GSH and γ-GC in one of the suppressor strains (AhpC V164G) to ensure that the mutation had not somehow restored synthesis of one of these thiol compounds; neither compound was detected (Fig. S1C). Thus, the mutant AhpC proteins are capable of restoring growth to the ΔgshA ΔtrxB strain in a manner that is independent of either GSH or γ-GC. Consequently, suppression must function via a different mechanism(s) than that mediated by previously isolated ahpC alleles. Interestingly, the new ahpC alleles could also restore growth to the ΔgshB ΔtrxB and Δgor ΔtrxB strains, whereas the suppressor mutations isolated from the former strains do not restore growth to the ΔgshA ΔtrxB strain (Table 1). Additionally, the Δgor ΔtrxB and ΔgshB ΔtrxB strains with these new ahpC alleles grow faster than the ΔgshA ΔtrxB strain with the same ahpC alleles (and the Δgor ΔtrxB strain grows slightly faster than the ΔgshB ΔtrxB strain with the same ahpC allele).
We asked whether the new AhpC mutant proteins may function by directly reducing oxidized glutaredoxins or thioredoxins. Marked grxA, trxA, or trxC deletions were transduced into the ΔgshA ΔtrxB suppressor strains harboring a trxB-complementing plasmid inducible with arabinose, and growth was assessed on rich agar media lacking arabinose. The ahpC mutants fell into different classes exhibiting different requirements for thioredoxins or glutaredoxins (Table 2). The growth of the suppressor strains containing AhpC S71F and AhpC S71F ΔE173 is drastically reduced or eliminated when either trxA or trxC is deleted, but not when the grxA gene is deleted. In contrast, the suppressor strain containing AhpC V164G does not require either the trxA or trxC gene for growth, but requires the grxA gene. Growth of the strain carrying the AhpC E171Ter suppressor is abolished when any one of the three genes trxA, trxC, and grxA is deleted. Finally, the suppressor strain containing the duplication of the region surrounding amino acid C165 does not appear to require trxA, trxC, or grxA because deletions of each individual gene did not affect colony size. These results suggest that the new ahpC class of suppressors is mechanistically heterogeneous because they supply electrons to the redox pathways via several distinct routes.
Table 2.
Requirement for glutaredoxin and thioredoxin in suppressors of thiol redox mutant strains
| Strain | Growth† | Growth with deletion† |
||
|---|---|---|---|---|
| ΔgrxA | ΔtrxA | ΔtrxC | ||
| Wild type | +++ | +++ | +++ | +++ |
| Δgor ΔtrxB ahpC* (+F38) | +++ | − | +++ | +++ |
| ΔgshB ΔtrxB ahpC* (+F38) | +++ | − | +++ | +++ |
| ΔgshA ΔtrxB ahpC (V164G) | ++ | − | ++ | ++ |
| ΔgshA ΔtrxB ahpC (E171Ter) | ++ | Very tiny | − | − |
| ΔgshA ΔtrxB ahpC (S71F) | + | + | − | ND |
| ΔgshA ΔtrxB ahpC (S71F, Δ E173) | ++ | ++ | − | − |
| ΔgshA ΔtrxB ahpC (G162–W169) | ++ | ++ | ++ | ++ |
†+++, grows similar to wild type (DHB4); ++, grows well but forms smaller colonies than wild type; +, forms tiny colonies after 1 day; Very tiny, forms good-sized colonies only after 2 days; −, does not grow; ND, growth not determined.
The ahpC suppressor mutations isolated previously require for their activity C165, the resolving cysteine of AhpC, but not the peroxidatic cysteine, C46 (4). The amino acid substitutions in AhpC caused by most of the suppressor mutations described here are close to C165, suggesting that they may use C165 to transfer electrons into the reductive pathways, analogously to that observed in the AhpC* enzyme. Plasmids expressing the new ahpC mutants and carrying either the C165S or the C46S mutations were analyzed for their ability to suppress the growth defect of the ΔgshA ΔtrxB ahpCF strain. In all cases tested, the C165S alteration eliminated suppressor activity (Table 3). However, the V164G and E171Ter mutants additionally showed some dependence on C46 because good-sized colonies of the C46S derivatives did not appear until after 2 days of growth. Because of the poor growth generally of these strains, we decided to do the same experiments in a strain background that grew better, a Δgor ΔtrxB strain. In this strain background, the mutants all showed dependence on C165 but not C46 for their suppressor activity (Table 3).
Table 3.
Requirement for cysteines by AhpC mutants for disulfide reductase activity
| AhpC protein | Growth of strain on NZ† |
|
|---|---|---|
| ΔgshA ΔtrxB ahpCF | Δgor ΔtrxB ahpCF | |
| Empty vector | − | − |
| Wild type | − | − |
| C46S | − | ++ |
| C165S | − | − |
| C46S, C165S | − | − |
| S71F | + | +++ |
| S71F, C46S | + | +++ |
| S71F, C165S | − | − |
| V164G | ++ | +++ |
| V164G, C46S | Very tiny | ++ |
| V164G, C165S | − | − |
| E171Ter | ++ | + |
| E171Ter, C46S | Very tiny | + |
| E171Ter, C165S | − | − |
†+++, grows similar to wild type (DHB4); ++, grows well but forms smaller colonies than wild type; +, forms tiny colonies after 1 day; Very tiny, forms good-sized colonies only after 2 days; −, does not grow.
Cytoplasmic Disulfide Bond Formation.
Wild-type E. coli strains do not ordinarily accumulate disulfide bonds in cytoplasmic proteins. However, we have shown that normally secreted proteins expressed without a leader peptide in the cytoplasm of a Δgor ΔtrxB ahpC* strain do accumulate largely in oxidized, correctly folded form (5). One such protein is alkaline phosphatase (PhoA), which contains two disulfide bonds that are essential for folding and catalytic activity. Disulfide bond formation occurs despite the presence of a pool of reduced GSH in this strain (4, 7). We examined whether the suppressor strains described here exhibited different properties in terms of their ability to make cytoplasmic disulfide bonds, and we examined both kinetics of disulfide bond formation and steady-state levels of oxidized proteins.
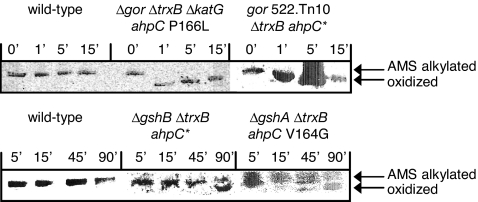
To evaluate the kinetics of disulfide bond formation in cytoplasmic PhoA, late-exponential phase cells were labeled with [35S]methionine for 1 min, at different times after the initiation of the chase. Disulfide bond formation was quenched with trichloroacetic acid, and free cysteines were reacted with 4-acetamido-4′-maleimidylstilbene-2,2′-disulfonic acid (AMS). PhoA was immunoprecipitated, and the oxidized and reduced forms of the protein were resolved by SDS/PAGE (Fig. 4). Rapid accumulation of oxidized PhoA occurred in the Δgor ΔtrxB supp strains irrespective of their ahpC suppressor allele. Specifically, the oxidation of PhoA in strains containing either the ahpC* or the ahpC P166L suppressor was complete in 1 min. In contrast, in the ΔgshB ΔtrxB ahpC* and ΔgshA ΔtrxB ahpC V164G strains, protein oxidation proceeded very slowly and was only ≈50% complete after 90 min. Even after adjusting for difference in growth rates (≈80 min for the Δgsh ΔtrxB supp strains compared with 45 min for the Δgor ΔtrxB ΔkatG ahpC P166L strain), disulfide bond formation in the Δgor ΔtrxB ΔkatG ahpC P166L strain was >20 times faster (katG encodes the primary catalase).
Fig. 4.
Kinetics of alkaline phosphatase oxidation. Oxidized and AMS-alkylated forms of PhoA were resolved by SDS/PAGE. Time points indicate the minutes after chase.
However, despite slow kinetics of disulfide bond formation in the ΔgshA ΔtrxB supp strains, these strains accumulated high levels of active PhoA at steady state in minimal media (Table 4). Active PhoA accumulated in the ΔgshA ΔtrxB strain containing two of the new ahpC suppressor alleles, ahpC V164G or ahpC E171Ter, at a level 120- to 160-fold higher than in the parental strain DHB4. The ΔgshB ΔtrxB ahpC* mutant strain displayed markedly lower PhoA activity (4-fold lower than ΔgshA ΔtrxB supp strains, although still ≈30-fold higher compared with the wild-type strain DHB4; Table 4 and Fig. S2C).
Table 4.
Yield of active, disulfide bond-containing proteins expressed in the cytoplasm
| Strain | vtPA (+DsbC) | Fab (1:2) | PhoA |
|---|---|---|---|
| Wild type | 1 ± 0.5 | 1 ± 0.3 | 1 ± 0.2 |
| Δgor ΔtrxB ahpC* | 119.4 ± 10.9 | 11.5 ± 0.6 | − |
| ΔgshB ΔtrxB ahpC* | 18.5 ± 2.8 | 5.0 ± 0.7 | 34.0 ± 2.0 |
| Δgor ΔtrxB ΔkatG ahpC P166L | 31.1 ± 10.2 | 5.7 ± 0.1 | 123.0 ± 5.0 |
| ΔgshA ΔtrxB ahpC V164G | 32.6 ± 5.3 | 12.1 ± 1.5 | 136.0 ± 13.0 |
| ΔgshA ΔtrxB ahpC E171Ter | 10.3 ± 9.4 | 27.6 ± 4.0 | 158 ± 3.0 |
Results shown are a summary of those in Fig. S2. Yields are relative to those in wild-type (DHB4). Bold entries show the best oxidation for a given substrate.
We also examined the influence of the different suppressor strains on other proteins having more complex disulfide bond patterns, vtPA and Fab antibody fragment. vtPA is a truncated form of the human tissue plasminogen activator containing nine disulfide bonds. Coexpression of the disulfide isomerase DsbC greatly enhances the folding of vtPA in the periplasm or in the cytoplasm (5, 8). Interestingly, the steady-state yields of PhoA did not correlate with the level of accumulation of active vtPA (Table 4 and Fig. S2A). Finally, we determined the cytoplasmic yield of Fab antibody fragment, which is a heterodimeric protein containing two disulfide bonds in each chain and a fifth interchain disulfide bond (9). In this case, the new class of ΔgshA ΔtrxB supp strains accumulated a substantially greater amount of functional Fab antibody (Table 4 and Fig. S2B). These results indicate that for proteins with complex patterns of disulfide bonds, optimal folding does not correlate with the rate of disulfide bond formation but is affected by whether the strains lack GSH or γ-GC and the nature of the suppressor mutations in ahpC.
Discussion
In this work, we have analyzed mutants of E. coli defective in both cytoplasmic disulfide-reducing pathways (the glutathione and thioredoxin pathways) and suppressors that restore growth to them. These studies have provided biological or biotechnological insights in three areas: they (i) shed additional light on the physiological functioning of the GSH pathway (and effects of the lack thereof); (ii) reveal surprising plasticity of the peroxidase AhpC because mutant versions of it provide several different means of restoring electron transfer to disulfide-reducing pathways; and (iii) give information on how protein disulfide bonds are formed in the cytoplasm because the different suppressor strains vary in their efficiency at introducing disulfide bonds into different proteins.
In the case of physiological implications, we confirm that a double-mutant strain that accumulates neither GSH nor its precursor γ-GC and that is defective in thioredoxin reduction (ΔgshA ΔtrxB) cannot grow. However, this growth defect can be overcome by ahpC mutations that alter AhpC so as to direct electron transfer to thioredoxins or glutaredoxins, obviating the need for GSH for survival. In contrast to the ΔgshA ΔtrxB strain, a double-mutant strain (ΔgshB ΔtrxB) defective in GSH biosynthesis but accumulating the GSH precursor γ-GC does grow, forming good-sized colonies after 2 days of incubation on rich media. Furthermore, the mutation ahpC*, which restores growth to a Δgor ΔtrxB strain by promoting the regeneration of reduced GSH, greatly enhances growth of the ΔgshB ΔtrxB strain where only γ-GC is present. We have shown that the presence of ahpC* in the ΔgshB ΔtrxB strain results in the accumulation of reduced γ-GC. Therefore, AhpC* is able to generate reduced γ-GC perhaps by a mechanism similar to that by which it generates reduced GSH. These results indicate that γ-GC can substitute to a significant extent for the redox properties of GSH in a background missing reduced thioredoxins.
The possibility that γ-GC can substitute for GSH in some instances is consistent with findings in other organisms. Halobacteria do not produce GSH, but accumulate γ-GC to millimolar levels (10). In addition, our results are similar to those obtained with Saccharomyces cerevisiae where there is evidence that γ-GC can substitute for GSH (11). Unlike E. coli, the S. cerevisiae gene gsh1, encoding γ-GC synthetase, which is homologous to gshA, is essential for growth under certain conditions and for protection against oxidative stress. However, a S. cerevisiae strain containing a deletion in gsh2, encoding GSH synthetase and the gene homologous to E. coli gshB, is viable and resistant to oxidative stress, indicating that in this organism γ-GC may partially replace GSH (12).
The plasticity of AhpC is indicated by our finding that the various ahpC suppressor alleles operate via distinct mechanisms. A previous study revealed only one class of ahpC mutant proteins, those that generate reduced GSH by cleaving gluthionylated glutaredoxins (4, 7). Here, we show that those proteins restore growth to a ΔgshB ΔtrxB strain and that the presence of one mutant protein in this class of mutants, ahpC*, results in the generation of reduced γ-GC in this strain, indicating that they are also able to reduce the GSH precursor γ-GC. In contrast, AhpC mutant proteins that suppress the growth defect of a ΔgshA ΔtrxB strain are functionally heterogeneous, passing their electrons into reductive pathways by different mechanisms, all of which are independent of GSH and its precursor γ-GC. Genetic studies show that mutant ahpC protein V164G requires (and AhpCE171Ter partially requires) glutaredoxin 1 to channel electrons into ribonucleotide reductase, AhpC S71F and AhpC E171Ter require thioredoxin 1 or 2, and AhpC (G162–W169 duplication) may use an entirely different substrate or use both glutaredoxins and thioredoxins. Although biochemical studies will be required to elaborate the details of their respective mechanisms of action, it is clear that the altered AhpCs change the specificity of AhpC so that its physiological role is changed (or expanded) from reduction of peroxides. The suppressor mutations described here may work directly through glutaredoxins (in the absence of GSH) and thioredoxins, may have broader specificity for such substrates, or may function through the intermediary of different redox-active small molecules or proteins.
To our knowledge, the emergence of such functional plasticity in the course of evolution under selective pressure is unusual. Although there are a few examples where mutagenesis approaches resulted in the generation of enzyme variants displaying extensive functional diversification (13, 14), they required structure-based information or mutagenesis strategies that cannot be accessed in the course of natural evolution. By contrast, the AhpC variants reported here arose spontaneously to compensate for the absence of the normal cellular thiol reduction pathways.
Thus, it appears that AhpC peroxiredoxin is a surprisingly malleable enzyme. The identified intermediaries in the activity of the AhpC mutant proteins described here are glutaredoxins and thioredoxins, proteins that contain the core protein fold that defines the thioredoxin superfamily. Given that previously isolated mutant AhpC proteins all exhibit increased activity toward glutathionylated glutaredoxins (4), it may be that changes in the degree or specificity of the reducing activity of AhpC toward thioredoxin family members is what allows the range of properties of mutant AhpC proteins described here. It is also possible that the suppressor amino acid alterations vary in their effects on the redox potential of AhpC. Peroxiredoxins from different organisms, such as AhpC (and the proteins that reduce them, such as AhpF) are evolutionarily linked to both the thioredoxin and glutathione/glutaredoxin pathways, depending on the organism (15). Isolation of mutants in AhpC that suppress defects in the cytoplasmic thiol redox pathways may thus reflect the evolution of AhpC from proteins in these pathways. The potential evolutionary significance of the suppressor mutations is amplified by the finding that some bacteria exhibit more than one homolog of AhpC, one version being very close to the E. coli AhpC and a second, more distant one, being altered in one or another of the same residues that is altered in our suppressors (M.J.F., D. Boyd, and J.B., unpublished results).
All of the AhpC suppressor mutations, including those previously isolated, cluster in two distinct regions of the protein, both of which are close to the region of the protein in the crystal structure that contains the disulfide bond (Fig. 3). The newly isolated S71F change clusters with the AhpC* (+F38) change and R119C isolated previously (3, 4). These changes reside near C46 and the active site of the wild-type protein. Mutations in this region may disrupt the redox activity of C46, the peroxidatic Cys of AhpC. Additionally, S71 is a highly conserved residue found in most peroxiredoxins, including those of eukaryotic origin, indicating that it may be an important residue for the wild-type peroxidase function of AhpC (15).
The other mutations described here (V164G, G162–W169 duplication, E171Ter, and ΔE173) and many of those isolated previously result in changes near or in the C-terminal extension, which contains C165. Further, these suppressors (as well as AhpC S71F and AhpC*) show a strong dependence on C165 for their activity. In wild-type AhpC, this region undergoes a large structural rearrangement during the redox cycle of the enzyme, becoming highly disorganized in the oxidized form of the protein (16). These suppressor mutations may open up this region of the protein, making C165 more accessible to external substrates. Similarly, the seven additional amino acids inserted by the G162–W169 duplication suppressor mutation may act by extending the region containing the C-terminal cysteine to the surface of the protein, thus making it more accessible to potential substrates.
Earlier we showed that a major consequence of the inactivation of both thioredoxin reductase and glutathione reductase is that the cytoplasm is rendered more oxidizing and allows efficient formation of disulfide bonds in normally secreted proteins expressed without a leader peptide (5, 6). Here, we report that this is the case also for suppressors of the double-mutant strains, ΔgshA ΔtrxB and ΔgshB ΔtrxB. However, the kinetics of disulfide bond formation, as monitored by the oxidation of PhoA, are substantially slower relative to the Δgor ΔtrxB ahpC* strain. This result reveals a specific role of GSH and γ-GC in the formation of disulfide bonds when the cytoplasm is rendered oxidizing. In an analogous manner, thioredoxin 1, which, like GSH, is normally a reductant, is also important for disulfide bond formation in a ΔtrxB strain (and presumably in a Δgor ΔtrxB ahpC* strain) (17). Additionally, the redox activity of thioredoxin 1 and other thioreodoxin family member proteins depends on both the target protein and the redox state of thioredoxin. For example, when thioredoxin is exported to the oxidizing periplasm, it efficiently forms disulfide bonds in substrate proteins (18, 19). Together, these findings raise the possibility that a mixed disulfide between a small molecule thiol, i.e., either GSH or γ-GC, and a protein such as thioredoxin 1 may be the optimal disulfide bond donor in the oxidative folding of PhoA in the cytoplasm. Nonetheless, as the data in Table 4 reveal, even though the kinetics of disulfide bond formation in the ΔgshA ΔtrxB and ΔgshB ΔtrxB strains are slow, appreciable amounts of active PhoA accumulate in all strains after overnight growth.
Unlike PhoA, Fab immunoglobulins and the vtPA variant of human tissue plasminogen activator contain five and nine disulfide bonds, respectively. These proteins exhibit more complex kinetics of oxidative folding that depend in part on disulfide bond isomerization. The yield of the active form of Fab or vtPA is strain-dependent and did not correlate with the kinetics of disulfide bond formation in PhoA. For instance, in the ΔgshA ΔtrxB ahpC E171Ter strain, Fab accumulates to the highest levels, whereas vtPA activity is strikingly lower. Likewise, the Δgor ΔtrxB ahpC* strain gave the highest level of vtPA, significantly greater than that found in the Δgor ΔtrxB ΔkatG ahpC P166L strain, indicating that the nature of the suppressor allele plays an important role in affecting the protein redox state in the cytoplasm. Our results suggest that by a careful testing of different strains with an oxidizing cytoplasm, one may be able to optimize yields of active protein for each substrate of interest.
Materials and Methods
Bacterial Strains and Plasmids.
Bacterial strains and plasmids were constructed by using standard genetic procedures. Details are described in SI Experimental Procedures and listed in Table S1. Oligonucleotide primers for site-directed mutagenesis used in this work are shown in Table S2.
Suppressor Strain Isolation.
Suppressor strains were isolated on plates lacking arabinose and DTT, as described in SI Experimental Procedures.
Kinetics of Alkaline Phosphatase Oxidation and Enzyme Assays.
The kinetics of disulfide bond formation were determined by pulse–chase experiments followed by blocking of free cysteines and separation of the oxidized and reduced forms by SDS/PAGE (for details, see SI Experimental Procedures). Enzyme assays for PhoA and vtPA and ELISAs for estimating the yield of Fab were performed by using published methods, as described in detail in SI Experimental Procedures.
Supplementary Material
Acknowledgments.
We thank Dr. Lluis Masip for preliminary experiments and Dr. Mimi Susskind (University of Southern California, Los Angeles) for the plasmid pMS421. This work was supported by National Institute of General Medical Sciences/National Institutes of Health Grants GM055090 and GM041883. J.B. is an American Cancer Society Professor.
Footnotes
Conflict of interest statement: A U.S. patent application disclosing aspects of this work has been filed jointly by the University of Texas and Harvard Medical School. A separate project on disulfide bond formation in J.B.'s laboratory is partially funded by New England Biolabs.
This article contains supporting information online at www.pnas.org/cgi/content/full/0801986105/DCSupplemental.
References
- 1.Ortenberg R, Gon S, Porat A, Beckwith J. Interactions of glutaredoxins, ribonucleotide reductase, and components of the DNA replication system of Escherichia coli. Proc Natl Acad Sci USA. 2004;101:7439–7444. doi: 10.1073/pnas.0401965101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gon S, et al. A novel regulatory mechanism couples deoxyribonucleotide synthesis and DNA replication in Escherichia coli. EMBO J. 2006;25:1137–1147. doi: 10.1038/sj.emboj.7600990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ritz D, Lim J, Reynolds CM, Poole LB, Beckwith J. Conversion of a peroxiredoxin into a disulfide reductase by a triplet repeat expansion. Science. 2001;294:158–160. doi: 10.1126/science.1063143. [DOI] [PubMed] [Google Scholar]
- 4.Yamamoto Y, et al. Mutant AhpC peroxiredoxins suppress thiol-disulfide redox deficiencies and acquire deglutathionylating activity. Mol Cell. 2008;29:36–45. doi: 10.1016/j.molcel.2007.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bessette PH, Åslund F, Beckwith J, Georgiou G. Efficient folding of proteins with multiple disulfide bonds in the Escherichia coli cytoplasm. Proc Natl Acad Sci USA. 1999;96:13703–13708. doi: 10.1073/pnas.96.24.13703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prinz WA, Åslund F, Holmgren A, Beckwith J. The role of the thioredoxin and glutaredoxin pathways in reducing protein disulfide bonds in the Escherichia coli cytoplasm. J Biol Chem. 1997;272:15661–15667. doi: 10.1074/jbc.272.25.15661. [DOI] [PubMed] [Google Scholar]
- 7.Masip L, Veeravalli K, Georgiou G. The many faces of glutathione in bacteria. Antioxid Redox Signal. 2006;8:753–762. doi: 10.1089/ars.2006.8.753. [DOI] [PubMed] [Google Scholar]
- 8.Qiu J, Swartz JR, Georgiou G. Expression of active human tissue-type plasminogen activator in Escherichia coli. Appl Environ Microbiol. 1998;64:4891–4896. doi: 10.1128/aem.64.12.4891-4896.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levy R, Weiss R, Chen G, Iverson BL, Georgiou G. Production of correctly folded Fab antibody fragment in the cytoplasm of Escherichia coli trxB gor mutants via the coexpression of molecular chaperones. Protein Expr Purif. 2001;23:338–347. doi: 10.1006/prep.2001.1520. [DOI] [PubMed] [Google Scholar]
- 10.Newton GL, Javor B. Gamma-glutamylcysteine and thiosulfate are the major low-molecular-weight thiols in halobacteria. J Bacteriol. 1985;161:438–441. doi: 10.1128/jb.161.1.438-441.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ohtake Y, Satou A, Yabuchi S. Isolation and characterization of glutathione biosynthesis-deficient mutation in Saccharomyces cerevisiae. Agric Biol Chem. 2007;54:3145–3150. [Google Scholar]
- 12.Grant CM, Maciver FH, Dawes IW. Glutathione is an essential metabolite required for resistance to oxidative stress in the yeast Saccharomyces cerevisiae. Curr Genet. 1996;29:511–515. doi: 10.1007/BF02426954. [DOI] [PubMed] [Google Scholar]
- 13.Yoshikuni Y, Ferrin TE, Keasling JD. Designed divergent evolution of enzyme function. Nature. 2006;440:1078–1082. doi: 10.1038/nature04607. [DOI] [PubMed] [Google Scholar]
- 14.Varadarajan N, Rodriguez S, Georgiou G, Iverson BL. Engineering a family of highly active and selective endopeptidases with programmed substrate specificities. Nat Chem Biol. 2008 doi: 10.1038/nchembio.80. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Copley SD, Novak WR, Babbitt PC. Divergence of function in the thioredoxin fold suprafamily: Evidence for evolution of peroxiredoxins from a thioredoxin-like ancestor. Biochemistry. 2004;43:13981–13995. doi: 10.1021/bi048947r. [DOI] [PubMed] [Google Scholar]
- 16.Poole LB. Bacterial defenses against oxidants: Mechanistic features of cysteine-based peroxidases and their flavoprotein reductases. Arch Biochem Biophys. 2005;433:240–254. doi: 10.1016/j.abb.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 17.Stewart EJ, Åslund F, Beckwith J. Disulfide bond formation in the Escherichia coli cytoplasm: An in vivo role reversal for the thioredoxins. EMBO J. 1998;17:5543–5550. doi: 10.1093/emboj/17.19.5543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Debarbieux L, Beckwith J. The reductive enzyme thioredoxin 1 acts as an oxidant when it is exported to the Escherichia coli periplasm. Proc Natl Acad Sci USA. 1998;95:10751–10756. doi: 10.1073/pnas.95.18.10751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Debarbieux L, Beckwith J. On the functional interchangeability, oxidant versus reductant, of members of the thioredoxin superfamily. J Bacteriol. 2000;182:723–727. doi: 10.1128/jb.182.3.723-727.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.