Abstract
ATP-dependent chromatin remodeling complexes are a notable group of epigenetic modifiers that use the energy of ATP hydrolysis to change the structure of chromatin, thereby altering its accessibility to nuclear factors. BAF250a (ARID1a) is a unique and defining subunit of the BAF chromatin remodeling complex with the potential to facilitate chromosome alterations critical during development. Our studies show that ablation of BAF250a in early mouse embryos results in developmental arrest (about embryonic day 6.5) and absence of the mesodermal layer, indicating its critical role in early germ-layer formation. Moreover, BAF250a deficiency compromises ES cell pluripotency, severely inhibits self-renewal, and promotes differentiation into primitive endoderm-like cells under normal feeder-free culture conditions. Interestingly, this phenotype can be partially rescued by the presence of embryonic fibroblast cells. DNA microarray, immunostaining, and RNA analyses revealed that BAF250a-mediated chromatin remodeling contributes to the proper expression of numerous genes involved in ES cell self-renewal, including Sox2, Utf1, and Oct4. Furthermore, the pluripotency defects in BAF250a mutant ES cells appear to be cell lineage-specific. For example, embryoid body-based analyses demonstrated that BAF250a-ablated stem cells are defective in differentiating into fully functional mesoderm-derived cardiomyocytes and adipocytes but are capable of differentiating into ectoderm-derived neurons. Our results suggest that BAF250a is a key component of the gene regulatory machinery in ES cells controlling self-renewal, differentiation, and cell lineage decisions.
Keywords: BAF250a (ARID1a), lineage commitment, mesoderm
Regulatory factors that control chromatin architecture (directly or indirectly) are potential key proteins for maintaining the pluripotent state or directing differentiation of early embryonic cells into distinct cell types (1). Such factors include ATP-dependent chromatin remodeling complexes that hydrolyze ATP to noncovalently restructure, mobilize, or eject nucleosomes to modulate transcription factor access to chromosomal DNA (2). Among the various members of the ATP-dependent chromatin remodeling superfamily is the SWI/SNF subfamily, consisting of two closely related SWI/SNF remodeling complexes BAF and PBAF in mammalian cells (3, 4).
BAF250a, a defining subunit of the BAF chromatin remodeling complex (5, 6), is a trithorax group (TrxG) protein (7). TrxG proteins were initially identified by their ability to antagonize the Polycomb group (PcG) proteins to maintain proper expression of many differentiation regulators during development (8). Interestingly, many PcG and TrxG proteins are chromatin modifying factors. Recent studies mapping the targets of PcG action in mouse and human ES cells suggest these proteins also play a role in sustaining a heritable epigenetic state uniquely associated with pluripotency (9, 10).
Given the influence of epigenetic factors in determining developmental potential (embryonic development, cellular differentiation, and nuclear reprogramming) and the documented role of SWI/SNF remodeling proteins in periimplantation development (11–15), we sought to test the relevance of this key BAF subunit in early developmental decisions in the mouse embryo. We also generated ES cells in which loss of BAF250a could be induced in vitro to assess the role of BAF in stem cell maintenance and differentiation. Here, we report that ablation of BAF250a results in early embryonic developmental arrest in mice. Our studies also indicate that BAF250a deficiency severely compromises pluripotency and self-renewal. Although loss of BAF250a in ES cells promotes differentiation under normal culture conditions, this effect appears to be cell lineage-specific. We have also identified genes regulated by BAF250a in ES cells, such as Sox2, Utf1, and Oct4, that partially explain the ES cell phenotype observed. Our results suggest that BAF250a-mediated chromatin remodeling plays a critical role in maintaining a particular chromatin configuration of its target genes that is essential for ES pluripotency and mesoderm formation.
Results
BAF250a Is Abundantly Expressed in Early-Stage Embryos and ES Cells.
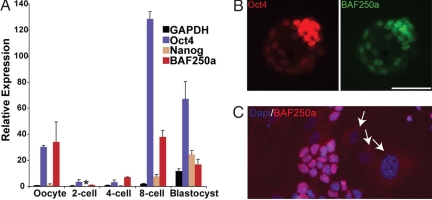
Quantitative RT-PCR and immunostaining assays were carried out to examine the expression of BAF250a in early-stage embryos and ES cells (Fig. 1). Like Brg1 (SMARCA4) (16), BAF250a appears to be a maternal transcript that is highly expressed in oocytes and the fertilized embryo before cell division (17) (Fig. 1A). Its expression is not detectable in embryos at the two-cell stage when general transcription is shut down. By the four- and eight-cell stages, BAF250a mRNA level increases sharply like Oct4, yet drops significantly at the blastocyst stage relative to the oocyte and eight-cell stages (Fig. 1A). Immunostaining of blastocyst stage embryos (Fig. 1B) confirms that BAF250a protein is very abundant in the preimplantation embryo. Similarly, the expression of BAF250a is abundant in ES cells at levels significantly higher than its expression in embryonic fibroblast cells (Fig. 1C). Thus, the expression pattern of BAF250a is consistent with a role in early embryogenesis and ES cell pluripotency.
Fig. 1.
Relative expression of BAF250a in early stage embryos and ES cells. (A) qRT-PCR analysis of relative expression of BAF250a, Oct4, Nanog, and GAPDH. (B) Immunostaining of BAF250a and Oct4 in blastocyst stage embryos. (C) Immunostaining of BAF250a expression in ES cells. *, not detectable expression; arrows, nuclei of fibroblast feeder cells. (Scale bar in B, 50 μm.)
Generation of BAF250a-Null Embryos and ES Cell Lines.
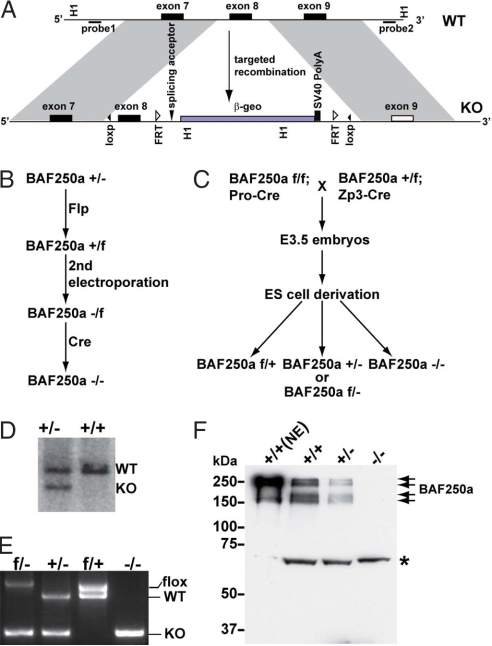
Unexpectedly, our initial attempts to generate BAF250a mutant mice either from gene trap insertions or standard targeted mutagenesis in ES cells were unsuccessful because of haploinsufficiency of BAF250a [supporting information (SI) Table S1 and Table S2, Fig. S1]. We therefore designed a knockout strategy to conditionally ablate BAF250a both in vivo and in vitro (Fig. 2A). We first generated BAF250a Het ES cell lines by gene targeting in E14 feeder-independent ES cells. After treatment with Flp, a reverted WT (rWT, f) allele is generated in which exon 8 of BAF250a is flanked by loxP sites.
Fig. 2.
Strategies to generate BAF250a conditional knockout embryos and ES cell lines. (A) Diagram of the strategy for homologous recombination. (B) Derivation of BAF250a homozygous ES cells from WT ES cells. (C) Derivation of BAF250a homozygous ES cells from embryos. See Materials and Methods for details. (D) Southern (E) RT-PCR and (F) Western blot analyses confirmed homologous recombination and indicated that BAF250a is absent in BAF250a homozygous ES cells. NE, nuclear extract; f, flox allele (reverted WT allele); H1, BamH I; Pro, protamine; *, a nonspecific band.
To generate BAF250a−/− ES cells in vitro, the rWT cells were electroporated a second time with the BAF250a knockout vector and selected in G418. Because of preferential retargeting of the first modified allele, only 1 in 15 of the colonies analyzed carried both targeted alleles (BAF250a−/f). To ablate BAF250a function in these cells, ligand-inducible Cre recombinase (Cre-ERT2) was introduced into the ROSA26 locus by gene targeting (18). Treatment of BAF250a−/f;Rosa26-CreERT2 cells with tamoxifen induced deletions between the two loxP sites in each targeted allele to produce cells lacking both copies of exon 8 of BAF250a (Fig. 2B). Removal of exon 8 is predicted to create a frameshift mutation and induce nonsense-mediated decay of the mutant transcript. Meanwhile, embryo-derived BAF250a-null ES cell lines were derived from BAF250a−/− blastocysts obtained from intercrosses of BAF250af/f;Pro-Cre males and BAF250af/+;Zp3-Cre females (Fig. 2C). Homologous recombination was identified by Southern blot analysis (Fig. 2D), PCR (Fig. 2E), and RT-PCR (data not shown), whereas the absence of BAF250a protein in BAF250a-null ES cell lines was confirmed by Western blot analyses (Fig. 2F and Fig. S2).
BAF250a Is Required for Early Embryo Development.
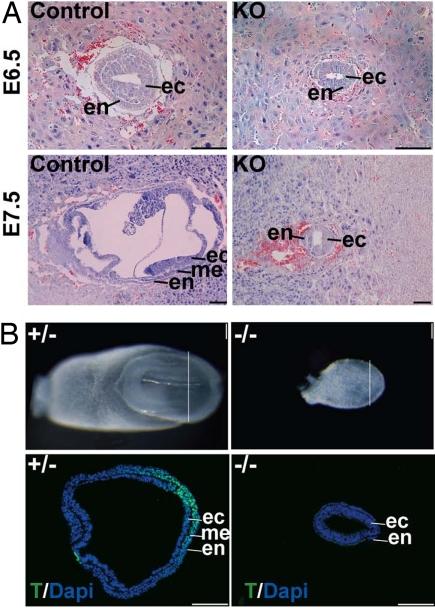
Embryos lacking BAF250a protein appear normal at embryonic day (E)3.5 blastocyst stage and formed trophectoderm and inner cell mass (ICM), as indicated by gross morphology and immunostaining against ICM marker Oct4 and trophectoderm marker Cdx2 (Fig. S3 A and B). Both mutant and control blastocysts developed normally to the late blastocyst stage after in vitro culture in KSOM medium (Fig. S3C). These data indicate that, whereas BAF250a is abundantly expressed in cleavage-stage embryos, zygotic BAF250a expression is not critical for preimplantation development. After implantation, however, BAF250a-null embryos arrest at E6.5 and fail to gastrulate. Sections through the deciduae at E6.5 and E7.5 reveal proper formation of the primitive endoderm and epiblast layers but a complete absence of mesoderm cells at this stage (Fig. 3). The absence of mesoderm was confirmed by an absence of Brachyury T immunostaining, a marker of the primitive streak, in serial sections of the whole embryo (Fig. 3B and data not shown). These results indicate that BAF250a is essential for proper germ-layer formation at gastrulation.
Fig. 3.
Ablation of BAF250a causes defects in early embryogenesis. (A) BAF250a−/− embryos failed to develop beyond E6.5, and to form mesoderm. (B) BAF250a−/− embryos failed to express Brachyury T. Embryos were serially sectioned and stained, and only one section is shown here. (Scale bar, 100 μm.)
BAF250a Is Required for Proper ES Cell Self-Renewal.
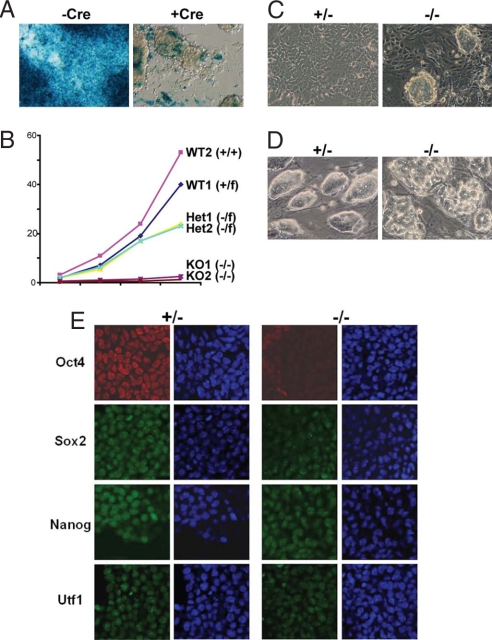
Factors maintaining the epiblast identity before gastrulation and directing germ-layer differentiation such as mesoderm formation very likely play important roles in ES cell pluripotency (1). Indeed, BAF250a homozygous mutant ES cells are unable to maintain a normal stem cell phenotype in culture. BAF250af/−;CreERT2 ES cell colonies after treatment with tamoxifen for 3 days displayed a highly abnormal morphology compared with control ES cells (Fig. 4A) and grew very slowly (Fig. 4B). After several passages, mutant cultures soon became dominated by cells with a primitive endoderm-like morphology. Mild trypsin treatment promoted maintenance of clusters of stem cells and reduced the degree of differentiation into primitive endoderm (Fig. 4C Right). We observed that many clusters became embryoid body (EB)-like structures and detached from the bottom of the gelatin-coated plates. After long-term culture, cells eventually adopted a uniform primitive endoderm-like morphology (data not shown), similar to the morphology and growth properties of extraembryonic endoderm (XEN) cells (19).
Fig. 4.
BAF250a ablation severely inhibits normal proliferation and promotes differentiation of ES cells. (A) Generation of BAF250 KO ES cells in vitro. LacZ staining indicates deletion of β-geo (KO allele) and conversion of BAF250a+/− to BAF250a−/− cells after tamoxifen induced Cre activation. Impaired growth and altered morphology are apparent within 76 h of drug addition. (B) Slow growth of BAF250a (−/−) ES cells. The plot represents the average of three separate experiments. (C) Lack of BAF250a promotes ES cell differentiation. The BAF250a Het (+/−, Left) ES cells grow and appear normal under feeder free culturing conditions, whereas Homo (−/−) ES cells form clusters, and differentiate into primitive endoderm-like cells (Right). (D) Morphology of embryo-derived BAF250a−/− ES cells at early passages with feeder cells. (E) Immunostaining of ES markers in BAF250a−/− ES cells.
To confirm that the phenotype observed in BAF250a−/− ES cells obtained in vitro was not due in part to additional mutations that may have arisen during the extensive manipulation of these cells in culture, we also derived BAF250a−/− ES cells from E3.5 BAF250a−/− embryos (Fig. 4D). The morphology and growth properties of embryo-derived BAF250a−/− ES cells cultured on feeders were visibly abnormal but less pronounced compared with the in vitro-derived mutant lines (Fig. 4, compare C and D). Interestingly, the severity of the phenotype appears to be ameliorated by the presence of feeder cells. When the embryo-derived BAF250a-null cells were cultured in the absence of MEFs, the cells displayed a phenotype very similar to the in vitro-derived feeder-free null ES cells (Fig. S4A). Conversely, when the conversion of the in vitro-derived BAF250a−/f to BAF250a−/− was performed in the presence of MEFs, the morphology and growth of BAF250−/− colonies recapitulated the less severe phenotype of embryo-derived feeder-dependent ES cell colonies (Fig. S4B). These data indicate that the cellular phenotype observed is due primarily to the loss of BAF250a and, furthermore, the presence of feeder cells can significantly suppress the phenotype of BAF250a mutant cells.
To assess the phenotype of BAF250a mutant cells at the molecular level, we performed fluorescence immunostaining using antibodies against the ES cell marker proteins. In vitro-derived mutant ES cells show a marked reduction in the expression of Oct4, Sox2, and Utf1 (Fig. 4E). Taken together, these results indicate that BAF250a mutant cells fail to properly maintain the expression of stem cell markers, and, consequently are prone to differentiate.
We next performed DNA microarray analyses to identify genes whose expression was affected by the ablation of BAF250a. Consistent with the defects in ES self-renewal manifested in BAF250a-null cells, some of the self-renewal genes were down-regulated in the mutant ES cells, such as Slc7a3 (≈11-fold), Fgf4 (≈10-fold), Sox2 and Utf1 (nearly 5-fold), and Oct4 (≈3-fold). Moreover, genes with known roles in early development and organogenesis were up-regulated in the BAF250a knockout ES cells, such as Gata4, Gata6, Cited1, Cited2, Cubn, Dab2, and Sox17 implicated in endoderm development and Tnt2, Myl3, and Ctgf, genes involved in cardiogenesis (Table S3). The down-regulation of self-renewal related genes and up-regulation of differentiation genes suggest that BAF250a is required for maintaining the ES cell state.
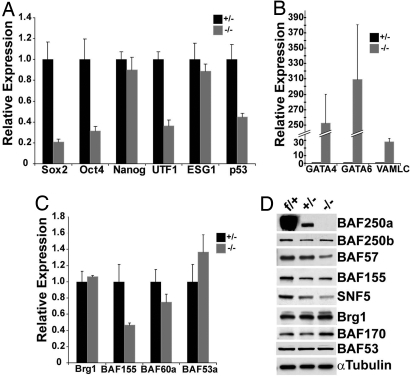
Quantitative RT-PCR analysis was used to further validate the expression changes in BAF250a mutant cells. Consistent with our expression microarray results, the expression of ES cell markers Oct4, Sox2, and Utf1 was decreased in BAF250a-deficient cells (Fig. 5A). In contrast, a dramatic increase in expression of Gata4 and Gata6 was observed in these cells (Fig. 5B). The increased expression of genes such as Gata4, Gata6, Sox17, Cited1, Lama1, Krt8, Krt18, Sparc, and Fabp3 (DNA microarray, Table S3) suggests that BAF250a deletion promotes the differentiation of ES cells into primitive endoderm- or XEN-like cells (19).
Fig. 5.
Genes affected in BAF250a deficient ES cells. (A) ES cell markers Oct4, Sox2, and Utf1 are down-regulated in BAF250a-deficient cells. (B) GATA-4, GATA-6, and VAMLC are up-regulated in BAF250a-deficient cells. (C) Expression of other BAF subunits in BAF250a-deficient cells. (D) Western blot analysis of BAF subunits in BAF250af/+, BAF250a+/−, and BAF250a−/− ES cells. VAMLC, ventricular alkaline myosin light chain.
One interesting question is whether the phenotype observed in BAF250a-deficient ES cells is due solely to the ablation of BAF250a subunit from the BAF complex or is compounded by changes in the composition of the protein complex as well. To address this issue, we examined the expression of other BAF subunits. At the transcriptional level, expression of BAF155 was decreased, whereas the expression of other subunits remained relatively unchanged (Fig. 5C). We then performed Western blot analysis to quantitate the expression of BAF subunit protein levels using newly embryo-derived ES cells at early passages for these experiments. Compared with the relatively stable expression of Brg1 and BAF250b, the expression of SNF5, BAF57, and BAF155 decreased in BAF250a-deficient ES cells, whereas BAF170 expression increased (Fig. 5D). Decreased BAF155 and increased BAF170 expression (20) suggests these early-passage embryo-derived BAF250a-deficient ES cells cultured on feeders have already acquired properties of differentiated cells, consistent with the up-regulation of endodermal cell markers in those in vitro-derived BAF250a−/− ES cells.
BAF250a Is Involved in Lineage-Dependent Differentiation of ES Cells.
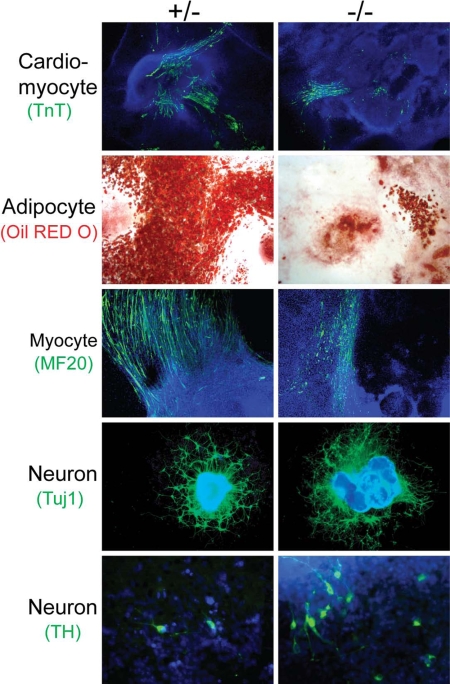
Given that BAF250a may be essential for ES self-renewal, and BAF250a−/− ES cells are prone to differentiate into endoderm-like cells, we next examined the differentiation potential of BAF250a mutant cells in EB differentiation assays in vitro. As shown in Table 1, similar proportions of BAF250a−/− and BAF250a+/− EBs contained Troponin T-positive cardiomyocytes. However, there are significantly fewer Troponin T-positive cardiomyocytes in each EB produced from knockout ES cells. Moreover, far fewer beating cardiomyocytes were observed in the knockout ES cell-derived EB cultures. Only 3% of EBs formed from knockout ES cells were beating compared with 68% of EBs originating from heterozygous cells (Table 1). This suggests that BAF250a is involved in the proper differentiation of functional cardiomyocytes, at least in our in vitro differentiation system.
Table 1.
Decreased differentiation into cardiomyocytes, adipocytes, and skeletal muscle cells in BAF250a knockout ES cells
| Cell type | Detection | Percentage of +/− EBs | Percentage of −/− EBs |
|---|---|---|---|
| Cardiomyocyte | TnT+ Staining | 70 ± 11 | 71 ± 7 |
| Beating | 68 ± 11 | 3 ± 1 | |
| Adipocyte | Oil red O+ | 57 ± 13 | 8 ± 4 |
| Skeletal muscle cells | MF20+ | 79 ± 13 | 72 ± 9 |
| Neuron | Tuj1+ | 97 ± 2 | 97 ± 4 |
| TH+ | 35 ± 1 | 96 ± 6 |
In addition to defective cardiomyocyte differentiation, a remarkable decrease in EB differentiation into adipocytes was also observed in knockout ES cells. Only 8% of EBs from knockout cells scored positive for adipocyte differentiation compared with 53% from heterozygous ES cells. Furthermore, there were dramatically fewer adipocytes evident in cultures of knockout EBs than in heterozygous EBs (Fig. 6 and Table 1). In addition, we detected a modest effect on the differentiation of BAF250a-null ES cells into skeletal muscle cells. In each EB, relatively fewer skeletal muscle cells were detected in mutant samples than in heterozygous controls.
Fig. 6.
Lineage-specific effect of BAF250a on ES cell differentiation. Differentiation potential of BAF250a−/− ES cells into cardiomyocytes, adipocytes, myocytes, and neurons. Cardiomyocyte, myocyte, and neuron formation are indicated by immunostaining against TnT, MF20, and Tuj1 and TH, respectively, whereas adipocyte formation is indicated by Oil red O staining.
In contrast, knockout ES cells appear to readily differentiate into neurons, with a similar percentage of EBs scoring Tubulin III-positive and an approximately equal number of neurons from each EB compared with heterozygous controls (Table 1 and Fig. 6). Furthermore, lack of BAF250a appears to promote tyrosine hydroxylase-positive (TH+) dopaminergic neuron formation. Ninety-six percent of EBs formed from knockout ES cells contained TH-positive cells compared with only 35% of EBs originating from heterozygous cells (Table 1). Moreover, on average, there were significantly more TH+ dopaminergic neurons in cultures of knockout EBs (≈42 per EB) than in heterozygous EBs (≈1 per EB). These data indicate that BAF250a is dispensable for the commitment of ES cells into neurons. Taken together, our results suggest that BAF250a function is important for the differentiation of mesodermal derivatives but appears dispensable for ectodermal lineages such as neuroectoderm.
Discussion
In this study, we have identified a requirement for the BAF250a protein, a unique and defining subunit of the chromatin remodeling complex BAF, both in mouse embryogenesis and in ES cells. Although inactivation of one allele resulted in late embryonic lethality, complete loss of BAF250a function caused developmental arrest around E6.5 without formation of a primitive streak and mesoderm. BAF250a is critical for the maintenance of ES cell self-renewal, and for lineage-specific differentiation of ES cells in vitro. Ablation of BAF250a in mouse ES cells led to abnormal cell morphology and impaired proliferation. Further analysis indicated that these changes were accompanied by diminished expression of numerous genes involved in ES cell self-renewal, including Oct4, Sox2, and Utf1, whereas the expression of primitive endoderm markers such as Gata4 and Gata6 was markedly increased.
BAF250a appears to be more important for the commitment of cell lineages of mesodermal origin. Extensive literature based on in vitro studies has linked SWI/SNF activity and the regulation of cellular differentiation, particularly in cell types of mesodermal origin (see review in ref. 2 and refs. therein), but these involve proteins present in both BAF and PBAF forms of the chromatin modification complexes. We have observed that BAF250a-deficient ES cells under feeder-free culture conditions eventually adopted a uniform primitive endoderm-like morphology. Our in vitro differentiation assays indicated that BAF250a-null ES cells are capable of adopting a neuronal fate. In fact, lack of BAF250a seems to promote TH+ dopaminergic neuron formation, suggesting potential opposing effects of BAF250a and neuronal-specific BAF subunits during neuron differentiation (21). In contrast, BAF250a-null ES cells are defective in differentiating into mesodermal lineages, including cardiomyocytes, adipocytes, and, to a lesser extent, skeletal muscles. Therefore, the lack of mesoderm in BAF250a-ablated embryos could be due at least in part to cell autonomous defects in lineage commitment in the epiblast. In this regard, it will be important to identify factors/signals regulated by BAF250a around E6 that are required for mesoderm formation.
Interestingly, the isolation of mutant ES cells in the presence and absence of feeder cells indicates that the severity of the ES cell phenotype is attenuated by the presence of embryonic fibroblast cells. Despite the complete feeder independence of the parental heterozygous ES line used to derive the mutant cells in vitro, it seems that the absence of feeder cells significantly impacts the self-renewal capacity and degree of spontaneous differentiation manifested in BAF250a-deficient cells. Presumably, one or more extracellular signaling pathways activated by coculture with feeder cells are involved in the suppression of the BAF250a mutant phenotype. The characterization of this signal(s) may help broaden our understanding of noncell autonomous factors required for stem cell maintenance and pluripotency.
This study has provided a useful clarification of the respective roles of the BAF and PBAF chromatin remodeling complexes during mammalian differentiation and development. Our results confirmed that BAF, unlike PBAF (22, 23), is essential for early embryogenesis and ES cell pluripotency. Both complexes share eight subunits, and several core constituents common to both, such as Brg1(SMARCA4), BAF155 (SMARCC1), and INI1 (SMARCB1), have been implicated in early embryogenesis and proliferation of the inner cell mass (from which ES cells are derived) (11–15). Our data show that the early lethality observed upon ablation of various core SWI/SNF chromatin remodeling subunits can be attributed chiefly to the loss of BAF250a containing BAF complexes, rather than the BAF180-associated PBAF complex, because BAF180 is dispensable for early development (22, 23).
BAF250a heterozygous mutant embryos consistently fail to complete normal embryonic development, necessitating a conditional approach to study the consequences of complete loss of function. To derive BAF250a null ES cell lines, we applied two complementary strategies. The first approach relied on the use of germ-line-specific Cre mouse lines to produce homozygous blastocysts from which ES cells were derived by using standard methods. The second consisted of an entirely in vitro approach in which both copies of the BAF250a gene were targeted sequentially in ES cells by using the same targeting vector (Fig. 2). The conditional allele generated by this strategy provides an effective means of eliminating gene function in a highly controlled manner after the activation of Cre-ERT2 with the ligand 4-hydroxytomoxifen (18). Our strategy is generally applicable to genes essential in ES cells, of which chromatin modifiers are strong candidates. The level of control endowed by the ligand-inducible Cre-ERT2 fusion will greatly facilitate future studies aimed at understanding the molecular changes over time that accompany the loss of BAF250a activity, and ultimately, the mechanistic basis of the phenotype observed in mutant cells.
Precisely how the lack of BAF250a leads to diminished pluripotency and spontaneous differentiation of ES cells into primitive endoderm needs further investigation. Nanog deficiency in ES cells also leads to primitive endoderm differentiation. Just before implantation, the ICM differentiates into primitive endoderm and primitive ectoderm (epiblast) around E4.5, and recent studies suggest that the balance between Nanog and Gata4/Gata6 expression is important for this lineage commitment (24). Our gene expression analyses indicate that primitive endoderm markers like Gata4, Gata6, Sox17, and Sparc are significantly up-regulated in mutant ES cells, confirming the primitive endoderm character of these cells. However, effects on Nanog expression are both subtle and variable in BAF250a mutant ES cells. In addition, BAF250a null ES cells derived in vitro continue to grow for many passages before converting completely to primitive endoderm-like cells. Finally, ES cell lines can be derived from BAF250a-deficient blastocysts, and the expression of SSEA1 but not Oct4 or Nanog appears to be down-regulated in embryo-derived mutant ES cells grown in the presence of feeders (Fig. S4 C and DRight), suggesting that inactivation of BAF250a in the embryo does not result in sudden and complete loss of pluripotency, as is the case for Nanog or Oct4. Both the temporal sequence of events involved in the loss of pluripotency and the ability of feeder cells to lessen this propensity to differentiate suggest that BAF250a does not directly regulate the transcription factors responsible for establishing pluripotency. Rather, we conclude that BAF250a influences the developmental potential and self-renewal/cell cycle properties of ES cells indirectly, in a manner that can be influenced by extracellular signaling pathways.
Materials and Methods
Generation of Conditional BAF250a Knockout Vector.
Two FRT and two loxp sites, together with a polylinker sequence, were engineered into a vector containing a promoterless β-geo trapping cassette derived from pGT1 (25) (Fig. 2A). DNA fragments ≈4 kb in length were PCR-amplified from genomic DNA 5′ and 3′ of exon 8 of BAF250a and inserted into the targeting vector (Fig. 2A). A 0.5-kb fragment containing exon 8 was PCR-amplified and inserted upstream of the β-geo trapping cassette.
Generation of BAF250a-Null ES Cell Lines in Vitro.
The BAF250a conditional knockout vector was linearized by NotI digestion and electroporated into E14 feeder-independent ES cells as described (26) to generate heterozygous ES lines after selection in G418. Selected heterozygous ES lines were transiently transfected with a FLP recombinase encoding plasmid (pCAGGsFLPe, from F. Stewart, University of Technology, Dresden, Germany), converting the initial knockout allele (BAF250a+/−, lacZ positive, G418 resistant) into a “wild-type” allele with two loxp sites flanking exon 8 [BAF250a+/f, reverted WT (rWT), lacZ negative, G418 sensitive] in 50–80% of the clones treated. Multiple independent rWT ES clones were then electroporated as described above with the original BAF250a conditional knockout vector and selected in G418. In the second round of targeting, 1/15 of the clones recovered were those in which the second WT allele was targeted (BAF250a−/f), confirmed by the presence of both the rWT allele (by genomic DNA PCR) and the second knockout allele (by RT-PCR as before). One of these clones was then stably transfected with the R26cre-ERT2-Pur plasmid (from J. Jonkeres, Netherlands Cancer Institute, Amsterdam) and selected in 1 μg/ml puromycin, to target a drug-inducible cre-mutant estrogen receptor fusion protein expression cassette into the ubiquitously expressed ROSA26 locus (18). Treatment of cells with 0.4 μM hydroxytamoxifen (OHT, Sigma) for 72 h induced Cre expression, promoting recombination between the loxP sites in both modified BAF250a alleles to convert the BAF250a−/f cells to BAF250a−/−. Because the recombination process was not 100% efficient (as monitored by X-Gal staining), OHT treated cells were then subcloned to produce pure BAF250a−/− cell lines.
Generation of Conditional BAF250a Knockout Embryos.
BAF250af/+ (rWT) ES cells were injected into blastocysts to generate BAF250af/+ mice that were intercrossed to generate BAF250af/f mice. These animals were then crossed with Protamine-Cre (Pro-Cre) transgenic mice to generate BAF250af/+;Pro-Cre double heterozygous mice. Female BAF250af/+;Pro-Cre mice were then crossed with BAF250af/f male mice to generate BAF250af/f;Pro-Cre mice. BAF250af/+;Zp3-Cre mice were obtained similarly by crossing BAF250af/f with Zp3-Cre mice. The BAF250af/f;Zp3-Cre mice were generated by crossing BAF250af/+;Zp3-Cre male mice with BAF250af/f female mice. BAF250a homozygous knockout embryos were obtained by crossing BAF250af/f;Pro-Cre male mice with BAF250af/f;Zp3-Cre or BAF250af/+;Zp3-Cre female mice.
Derivation of BAF250a-Null ES Cell Lines from Blastocysts.
E3.5 blastocysts were flushed out of the uterus of pregnant BAF250af/+;Zp3-Cre female mice mated with BAF250af/f;Pro-Cre male mice, and procedures for derivation of BAF250a null ES cell lines were as described (27).
Southern and Western Blot Analyses.
For Southern blot analysis, BamH I digestion produces a 10-kb fragment for WT and 7-kb fragment for mutant alleles using a PCR-amplified genomic probe downstream of the 3′ homology arm (as indicated in Fig. 2A). For Western blot analysis, cells were lysed either in 1× SDS loading buffer or in 0.25% Triton X-100; 50 mM Tris·HCl, pH 7.4; 150 mM NaCl; 1 mM EDTA with protease inhibitors (Roche). Rabbit antibodies against BAF250a, BAF250b, Brg-1, BAF155, BAF170, BAF53, BAF57, and SNF5 were generously provided by Weidong Wang (National Institutes of Health, Bethesda, MD). Mouse anti-Oct3/4 antibody was from Santa Cruz Biotechnology, and mouse anti-α-tubulin and -β-actin antibodies were from Sigma.
Histological Analysis.
Paraffin embedding, section preparation, deparaffinization, and H&E staining were carried out by using standard histological procedures.
Brachyury-T Immunostaining.
Embryos, fixed in 4% paraformaldehyde at 4°C overnight, were imbedded in OCT, and 8-μm cryosections were collected on a cryotome. Sections were blocked with 10% donkey serum in PBS buffer containing 0.1% Tween 20 for 1 h at 37°C and incubated with goat anti-Brachyury T (N-19) (Santa Cruz Biotechnology) at 37°C for 1 h. Then sections were washed three times with PBS buffer with 0.1% Tween 20 before being incubated with a donkey anti-goat Alexa 488-conjugated secondary antibody at 37°C for 1 h. After washes, section slides were mounted and analyzed by fluorescent microscopy.
Immunocytochemistry.
For antibody staining, ES cells were seeded on Permanox chamber slides (Nunc) and fixed 48–72 h later in −20°C methanol (Nanog, Sox2, and Utf1) or 4% paraformaldehyde (Oct 4). Fixed cells were incubated with polyclonal antibodies against Oct3/4 (Santa Cruz Biotechnology), Sox2 (Chemicon), Nanog (Abcam), Utf1 (Abcam), and SSEA1 (Developmental Studies Hybridoma Bank) at 1:100, 1:500, 1:150, 1:300, and 1:200 dilutions, respectively, followed by incubation with anti-goat IgG Alexa 594 (Oct 3/4), anti-rabbit IgG Alexa 488 (Sox2, Nanog, and Utf1), or anti-mouse IgM Alexa 488 (SSEA1) conjugates (Molecular Probes). Blastocysts were fixed in 4% paraformaldehyde and stained with mouse anti-Oct3/4 (Santa Cruz Biotechnology) and mouse anti-Cdx2 (BioGenex) at 1:100 and 1:20 dilutions, respectively. Nuclei were stained with Hoechst 33452 (Sigma) or Dapi (Dako).
Quantitative RT-PCR.
Total RNA was isolated from proliferating WT (BAF250a+/+, BAF250a+/f), heterozygous (BAF250a+/−, BAF250a−/f), and homozygous (BAF250a−/−) ES cells by using TRIzol (Invitrogen) and RNeasy Mini kit (Qiagen), as described (23). Quantitative RT-PCR was as described (23).
ES Cell Differentiation Assays.
EB-based differentiation assays were performed to assess the function of BAF250a in lineage-specific differentiation. Because of the slow growth of BAF250a-null ES cells, 1,500 homozygous ES cells were seeded in one hanging drop compared with 500 heterozygous ES cells, and the procedures for inducing different cell types were as described (28). Typically, >50 EBs were scored for each cell type, and at least two independent experiments were performed for each cell type.
Supplementary Material
Acknowledgments.
We thank J. Song, L. Caron, A. Sharov, T. Tenzen, C. Cowan, and X. Jiang for technical assistance and W. Wang for providing antibodies against BAF250a. This work was supported in part by an internal grant from Massachusetts General Hospital (Z.W.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The DNA microarray data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE10553).
This article contains supporting information online at www.pnas.org/cgi/content/full/0801802105/DCSupplemental.
References
- 1.Surani MA, Hayashi K, Hajkova P. Genetic and epigenetic regulators of pluripotency. Cell. 2007;128:747–762. doi: 10.1016/j.cell.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 2.de la Serna IL, Ohkawa Y, Imbalzano AN. Chromatin remodelling in mammalian differentiation: lessons from ATP-dependent remodellers. Nat Rev Genet. 2006;7:461–473. doi: 10.1038/nrg1882. [DOI] [PubMed] [Google Scholar]
- 3.Wang W. The SWI/SNF family of ATP-dependent chromatin remodelers: similar mechanisms for diverse functions. Curr Top Microbiol Immunol. 2003;274:143–169. doi: 10.1007/978-3-642-55747-7_6. [DOI] [PubMed] [Google Scholar]
- 4.Roberts CW, Orkin SH. The SWI/SNF complex–chromatin and cancer. Nat Rev Cancer. 2004;4:133–142. doi: 10.1038/nrc1273. [DOI] [PubMed] [Google Scholar]
- 5.Dallas PB, et al. The human SWI-SNF complex protein p270 is an ARID family member with non-sequence-specific DNA binding activity. Mol Cell Biol. 2000;20:3137–3146. doi: 10.1128/mcb.20.9.3137-3146.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nie Z, et al. A specificity and targeting subunit of a human SWI/SNF family-related chromatin-remodeling complex. Mol Cell Biol. 2000;20:8879–8888. doi: 10.1128/mcb.20.23.8879-8888.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vazquez M, Moore L, Kennison JA. The trithorax group gene osa encodes an ARID-domain protein that genetically interacts with the brahma chromatin-remodeling factor to regulate transcription. Development. 1999;126:733–742. doi: 10.1242/dev.126.4.733. [DOI] [PubMed] [Google Scholar]
- 8.Grimaud C, Negre N, Cavalli G. From genetics to epigenetics: the tale of Polycomb group and trithorax group genes. Chromosome Res. 2006;14:363–375. doi: 10.1007/s10577-006-1069-y. [DOI] [PubMed] [Google Scholar]
- 9.Boyer LA, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 10.Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 11.Bultman S, et al. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol Cell. 2000;6:1287–1295. doi: 10.1016/s1097-2765(00)00127-1. [DOI] [PubMed] [Google Scholar]
- 12.Roberts CW, et al. Haploinsufficiency of Snf5 (integrase interactor 1) predisposes to malignant rhabdoid tumors in mice. Proc Natl Acad Sci USA. 2000;97:13796–13800. doi: 10.1073/pnas.250492697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klochendler-Yeivin A, et al. The murine SNF5/INI1 chromatin remodeling factor is essential for embryonic development and tumor suppression. EMBO Rep. 2000;1:500–506. doi: 10.1093/embo-reports/kvd129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim JK, et al. Srg3, a mouse homolog of yeast SWI3, is essential for early embryogenesis and involved in brain development. Mol Cell Biol. 2001;21:7787–7795. doi: 10.1128/MCB.21.22.7787-7795.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guidi CJ, et al. Disruption of Ini1 leads to peri-implantation lethality and tumorigenesis in mice. Mol Cell Biol. 2001;21:3598–3603. doi: 10.1128/MCB.21.10.3598-3603.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bultman SJ, et al. Maternal BRG1 regulates zygotic genome activation in the mouse. Genes Dev. 2006;20:1744–1754. doi: 10.1101/gad.1435106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sharov AA, et al. Transcriptome analysis of mouse stem cells and early embryos. PLoS Biol. 2003;1:410–419. doi: 10.1371/journal.pbio.0000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vooijs M, Jonkers J, Berns A. A highly efficient ligand-regulated Cre recombinase mouse line shows that LoxP recombination is position dependent. EMBO Rep. 2001;2:292–297. doi: 10.1093/embo-reports/kve064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kunath T, et al. Imprinted X-inactivation in extra-embryonic endoderm cell lines from mouse blastocysts. Development. 2005;132:1649–1661. doi: 10.1242/dev.01715. [DOI] [PubMed] [Google Scholar]
- 20.Yan Z, et al. The BAF250b-associated SWI/SNF chromatin-remodeling complex is required for the maintenance of undifferentiated mouse embryonic stem cells. Stem Cells. 2008 doi: 10.1634/stemcells.2007-0846. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu JI, et al. Regulation of dendritic development by neuron-specific chromatin remodeling complexes. Neuron. 2007;56:94–108. doi: 10.1016/j.neuron.2007.08.021. [DOI] [PubMed] [Google Scholar]
- 22.Huang X, et al. Coronary development is regulated by ATP-dependent SWI/SNF chromatin remodeling component BAF180. Dev Biol. 2008 doi: 10.1016/j.ydbio.2008.04.020. in press. [DOI] [PubMed] [Google Scholar]
- 23.Wang Z, et al. Polybromo protein BAF180 functions in mammalian cardiac chamber maturation. Genes Dev. 2004;18:3106–3116. doi: 10.1101/gad.1238104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boyer LA, Mathur D, Jaenisch R. Molecular control of pluripotency. Curr Opin Genet Dev. 2006;16:455–462. doi: 10.1016/j.gde.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 25.Wilson V, et al. The T gene is necessary for normal mesodermal morphogenetic cell movements during gastrulation. Development. 1995;121:877–886. doi: 10.1242/dev.121.3.877. [DOI] [PubMed] [Google Scholar]
- 26.Skarnes WC. Gene trapping methods for the identification and functional analysis of cell surface proteins in mice. Methods Enzymol. 2000;328:592–615. doi: 10.1016/s0076-6879(00)28420-6. [DOI] [PubMed] [Google Scholar]
- 27.Nagy A, Gertsenstein M, Vintersten K, Behringer R. In: Manipulating the Mouse Embryo. Nagy A, Gertsenstein M, Vintersten K, Behringer R, editors. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2003. pp. 359–398. [Google Scholar]
- 28.Caron L, et al. A new role for the oncogenic high-mobility group A2 transcription factor in myogenesis of embryonic stem cells. Oncogene. 2005;24:6281–6291. doi: 10.1038/sj.onc.1208781. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.